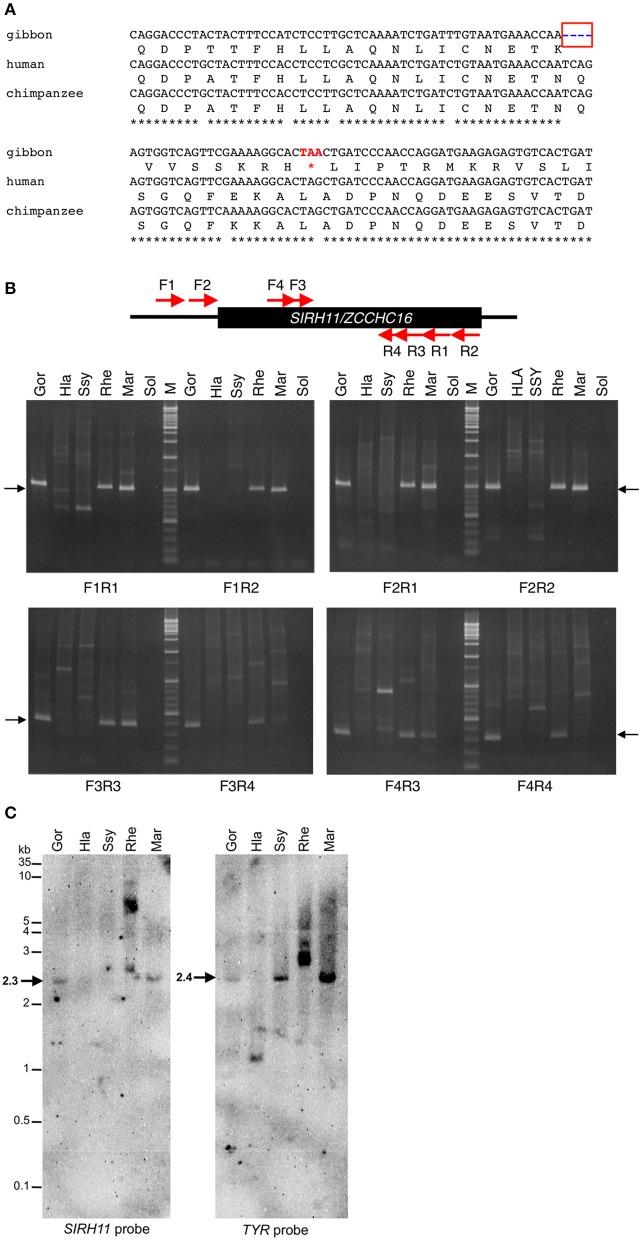
Figure 2.
(A) Nonsense mutation in gibbon SIRH11/ZCCHC16. The four bp deletion (blue in a red box) in gibbon leads to a nonsense mutation (red). Note that there is a G to A transition (DNA polymorphism) in a stop codon of gibbon (TAA) compared with human/chimpanzee and other primates (TAG). (B) PCR analysis of gibbon SIRH11/ZCCHC16. Upper panel shows the schematic representation of primer design. Lower panel shows agarose gel electrophoresis profile in each primer set. The arrows represent expected band size. M, 100 bp and 1 kb ladder; Gor, gorilla; Hla, white-handed gibbon; Ssy, siamang; Rhe, rhesus macaque; Mar, common marmoset; Sol, solvent only (no DNA). (C) Southern blot analysis of Hla and Ssy. Left and right panels show the result of hybridization using SIRH11 and TYR probes, respectively. The arrows indicate expected band size. Gor, gorilla; Hla, white-handed gibbon; Ssy, siamang; Rhe, rhesus macaque; Mar, common marmoset. (D) Amino acid sequence alignment of Chiroptera SIRH11/ZCCHC16. The blue asterisks in a red box indicate a common nonsense mutation in megachiroptera. The asterisks, colons and periods below the amino acids indicate identical, strongly and weakly similar residues among six species, respectively. (E) DNA alignments around the common TAA nonsense mutation (red).