Figure 3.
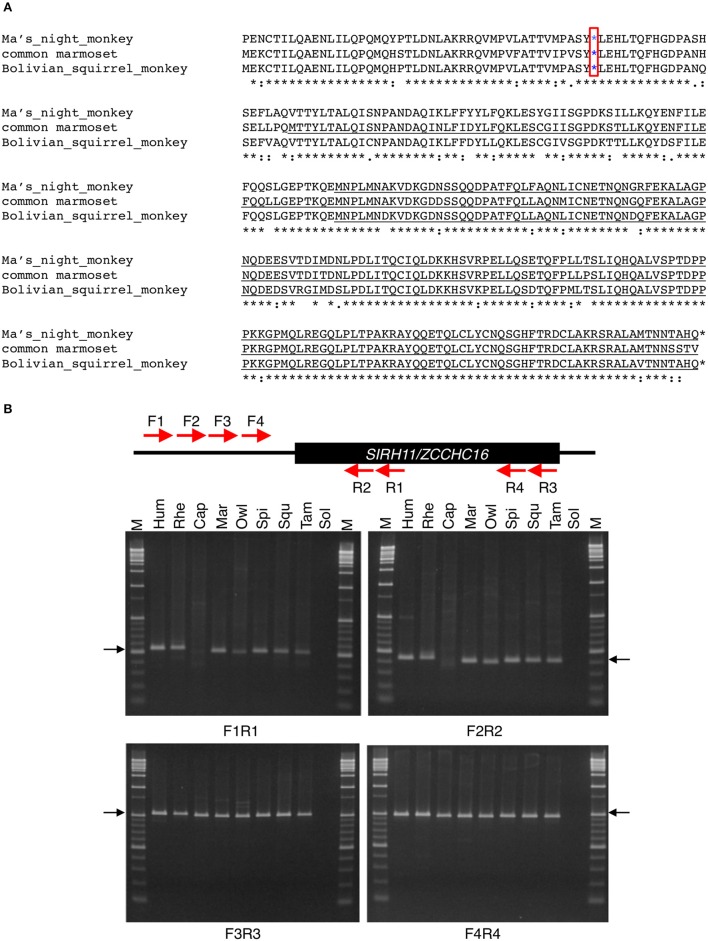
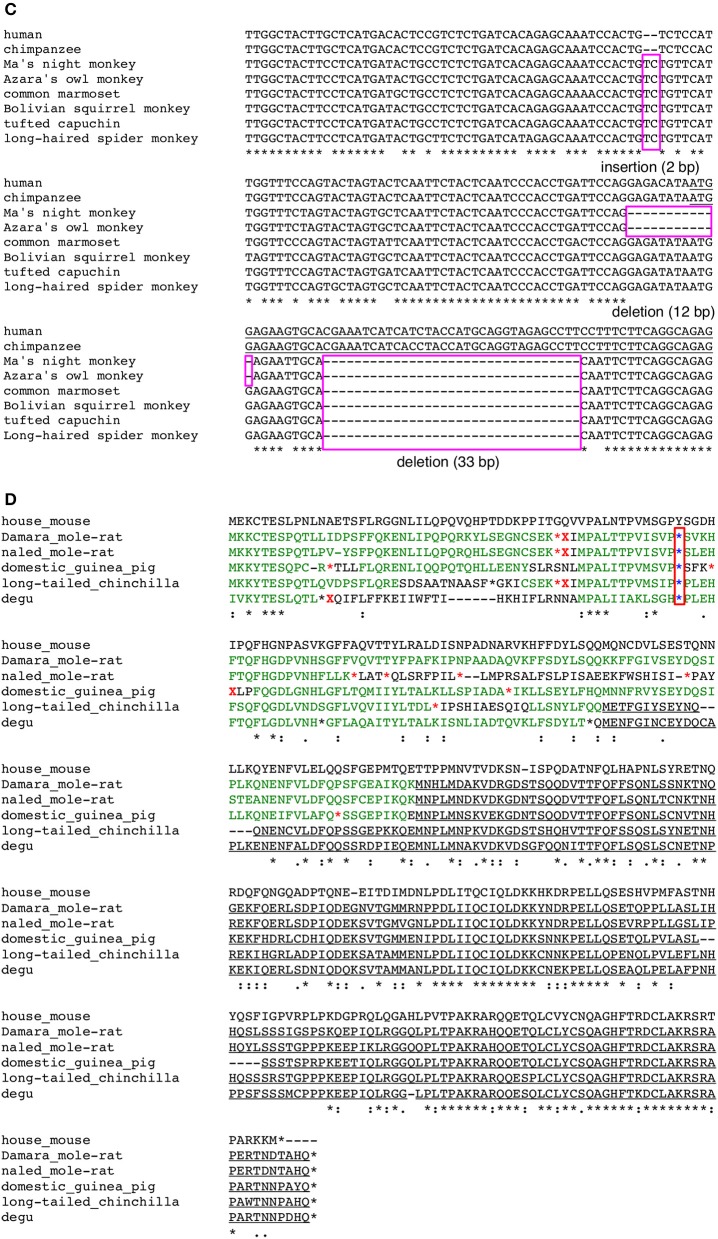
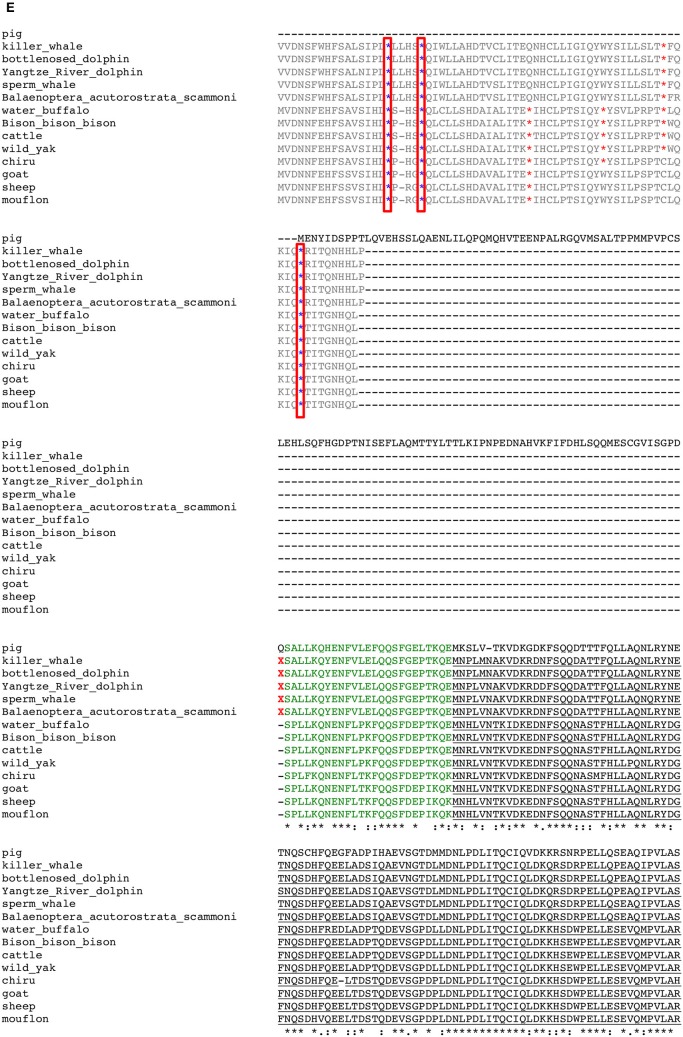
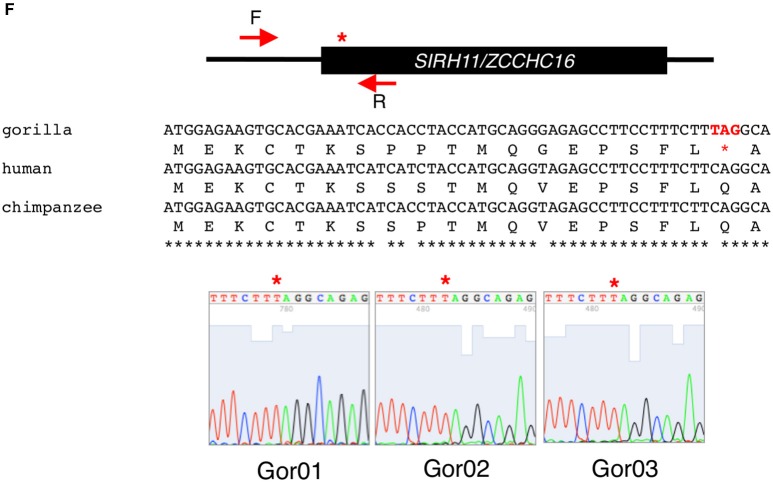
(A) Amino acid sequence alignment of Platyrrhihi SIRH11/ZCCHC16. The blue asterisks indicate common nonsense mutations in Platyrrhihi. The underlined sequences indicate the putative short ORFs starting from a next Met codon. The asterisks, colons and periods below the amino acids indicate identical, strongly and weakly similar residues among three species, respectively. (B) PCR analysis of Platyrrhihi SIRH11/ZCCHC16. Upper panel shows the schematic representation of the primer design. Lower panel shows agarose gel electrophoresis profile in each primer set. The arrows represent expected band size. M, 100 bp and 1 kb ladder; Hum, human; Rhe, rhesus macaque; Cap, tufted capuchin; Mar, common marmoset; Owl, the Azara's owl monkey; Spi, long-haired spider monkey; Squ, common squirrel monkey; Tam, cotton-top tamarin; Sol, solvent only (no DNA). (C) DNA sequence analysis of Platyrrhihi SIRH11/ZCCHC16. DNA sequences of Azara's owl monkey, common marmoset, tufted capuchin, and long-haired spider monkey determined by our own experiments are also shown. Magenta boxes show lineage specific insertion or deletion. The underlined letters indicate the SIRH11/ZCCHC16 ORF. (D) Amino acid sequence alignment of Hystricognathi SIRH11/ZCCHC16. The similar sequences among five Hystricognathi species are expressed in green. The red asterisks and Xs indicate the sites of ORF termination and frameshift, respectively. The blue asterisks in a red box indicate a common nonsense mutation in Hystricognathi. The underlined sequences indicate the putative short ORFs starting from a next Met codon. The asterisks, colons and periods below the amino acids indicate identical, strongly and weakly similar residues among six species, respectively. The house mouse sequence is used as a reference. (E) Amino acid sequence alignment of Cetartiodactyla SIRH11/ZCCHC16. The blue asterisks in red boxes indicate common nonsense mutations in Cetartiodactyla. The red asterisks and Xs indicate the sites of ORF termination and frameshift, respectively. The underlined sequences indicate the putative short ORFs starting from a next Met codon. The similar sequences among 14 Cetartiodactyla species are shown in green. The asterisks, colons and periods below the amino acids indicate identical, strongly and weakly similar residues among 14 species, respectively. The pig sequence is used as a reference. (F) Sequence analysis of gorilla SIRH11/ZCCHC16. Upper panel shows the schematic representation of the primer design and nonsense mutation site (red asterisk) in gorilla SIRH11/ZCCHC16. Middle panel shows the sequence comparison between gorilla, human, and chimpanzee. Lower panel represents the sequence results of three individuals.