Abstract
Chronic hepatitis B virus (HBV) infection remains a major health problem, with more than 240 million people chronically infected worldwide and potentially 650000 deaths per year due to advanced liver diseases including liver cirrhosis and hepatocellular carcinoma (HCC). HBV-X protein (HBx) contributes to the biology and pathogenesis of HBV via stimulating virus replication or altering host gene expression related to HCC. The HBV X region contains only 465 bp encoding the 16.5 kDa HBx protein, which also contains several critical cis-elements such as enhancer II, the core promoter and the microRNA-binding region. Thus, mutations in this region may affect not only the HBx open reading frame but also the overlapped cis-elements. Recently, several types of HBx mutations significantly associated with clinical severity have been described, although the functional mechanism in most of these cases remains unsolved. This review article will mainly focus on the HBx mutations proven to be significantly related to clinical severity via epidemiological studies.
Keywords: Hepatitis B virus infection, Hepatitis B virus-X protein mutation, Hepatocellular carcinoma, Clinical severity
Core tip: Of hepatitis B virus (HBV)-X protein (HBx) mutations related to clinical severity, the A1762T/G1764A BCP mutation is one of the most frequently encountered HBx mutations and plays a significant role in liver disease progression in chronic patients with HBV infections. It also further contributes to disease progression by inducing mutations of other HBx mutations related to clinical severity, such as G1386A/C (V5M/L), C1653T (H94Y), T1753V (I127V) and HBx C-terminal deletion or insertion. Moreover, T1753V (I127L,T,N,S) and C1653T (H94Y) mutations in the enhancer II/BCP region and A1383C, G1386A/C (V5M/L) and C1485T (P38S) in the negative regulation domain are significantly related to severe types of liver diseases, including hepatocellular carcinoma. Furthermore, deletions or insertions affecting the C-terminal region of HBx may also contribute to the severity of the clinical outcome in chronic patients. In addition, our recent study indicated that a novel mutation type (X8Del) composed of an 8-bp deletion in the C-terminal region of the HBx could contribute to occult HBV infection in vaccinated Korean individuals via a reduced secretion of HBsAg and virions. Therefore, several distinct types of HBx mutations may contribute to HBV pathogenesis by regulating HBV replication or host genes related to cell homeostasis.
INTRODUCTION
Chronic hepatitis B virus (HBV) infection remains a major health problem with more than 240 million people chronically infected worldwide, which potentially causes 650000 deaths per year due to advanced liver diseases including liver cirrhosis and hepatocellular carcinoma (HCC), particularly in endemic areas such as China and South Korea[1,2]. It is generally accepted that HBV infection accounts for approximately 50% of the HCC cases worldwide and even 80%-90% in highly endemic areas[1].
HBV is an enveloped Hepadnavirus belonging to the Hepadnaviridae family, with an incomplete double-stranded DNA genome of approximately 3.2 kb in length with four overlapping open reading frames (ORFs) encoding the polymerase (P), core (C), surface antigen (S), and X protein[3]. The S gene encodes a family of surface antigen polypeptides embedded within the viral envelope, which is a major target for diagnosis and protective vaccines. The C gene encodes the core antigen, which forms the nucleocapsid, within which reverse transcription of pre-genomic RNA occurs. The P gene encodes the virus reverse transcriptase, which also has RNase H and DNA polymerase activities[4-6]. Transcription of HBV proteins is controlled under four promoters (preS1, preS2, core and X) and two enhancers (EnhI and EnhII) in the viral genome, which overlap with those ORFs. Because it contains a polymerase without proofreading activity and uses an RNA intermediate (pgRNA) during its replication, the HBV genome has a higher mutation ratio than other DNA viruses[7-11]. Moreover, host immune pressures and interventions such as antiviral drugs and vaccines make the viral mutations more complicated[12-18].
Based on an intergroup divergence of > 8% in its complete genome sequence, the HBV strains are classified into eight genotypes, which are designated A-H, with a distinct ethnic and geographical distribution[1,19-21]. Different genotypes have distinct geographical distributions and usually induce various clinical outcomes. For instance, genotype C, the most prevalent genotype in Asia, is more prone to mutations and is associated with more severe liver diseases and lower antiviral responses compared with genotype B[3,22,23]. In particular, genotype C2 is reportedly responsible for the most chronic infections in South Korea. Indeed, several types of HBV mutations that are never or rarely encountered in other areas have been found in South Korea and have been proven through molecular epidemiologic or functional studies to be related to disease progression in chronic patients[24-44].
HBV X PROTEIN STRUCTURE AND FUNCTION
The HBV X protein (HBx) is a multifunctional nonstructural protein that contributes to HBV biology and pathogenesis by stimulating virus replication or altering host gene expression related to HCC. HBx contains only 465 bp encoding the 16.5 kDa protein, which also contains several critical cis-elements such as EnhII, the core promoter and the microRNA-binding region[45-47] (Figure 1).
Figure 1.
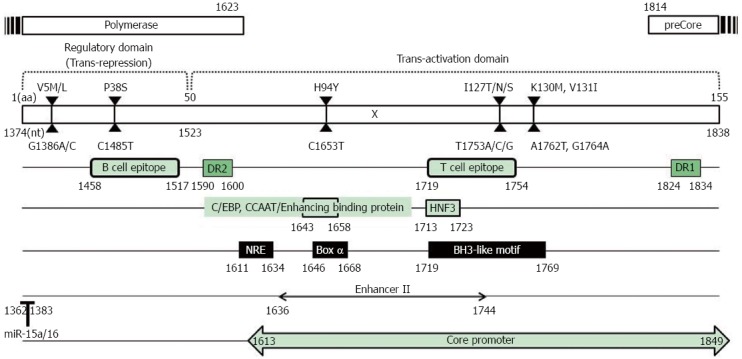
Hepatitis B virus-X protein genome structure. The HBV X region contains 465 bp (nt 1374 to 1838) encoding the 16.5 kDa HBx protein composed of 154 aa, which also contains several critical cis-elements such as EnhII (nt 1636 to 1744), the core promoter (nt 1613 to 1849) and the microRNA-binding region (nt 1362 to 1383). Thus, HBx mutations affected not only the HBx open reading frame but also the overlapped cis-elements. HBx: Hepatitis B virus-X protein.
HBx plays a significant role in sustained HBV replication, which is a major risk factor for HCC development via proteasome inhibition[48,49], transactivation of HBV enhancer or promoters[50], autophagy induction[51,52], or polymerase activation by Ca2+-dependent signaling[53-55]. HBx can also regulate HBV replication through epigenetic modifications, by being recruited onto the viral minichromosome in the nuclei of infected hepatocytes along with cellular histone acetyltransferases such as CREB-binding protein (CBP)/p300[56,57] and histone deacetylases such as HDAC1 and hSirt1[58]. HBx can help establish and maintain chronic infection by altering the patterns of host innate immunity, which causes the development and progression of chronic liver diseases in the absence of virus elimination[59,60]. HBx blocks apoptotic signaling and activates signaling pathways (such as NF-κB and PI3K) that override apoptotic signals from extrinsic ligands such as Fas or TNF-alpha[61,62]. HBx also plays an important role in hepatocarcinogenesis by inactivating the tumor suppressor p53[63], promoting Rb inactivation by phosphorylation[64], and compromising DNA repair mechanisms[65]. Consequently, mutations in the HBx ORF region may affect not only the HBx ORF and the overlapped cis-elements but also its binding capacity for host proteins. Recently, several types of HBx mutations significantly associated with clinical severity have been described mainly from chronic patients infected with genotype C[28,31,39,66-83], although the functional mechanism in most of these cases remains unsolved. This review article will mainly focus on the HBx mutations that have been proven to be significantly related to clinical severity via epidemiological studies (Table 1).
Table 1.
Mutations in the hepatitis B virus X region as related to clinical severity
| Type of mutation | Mutations |
Genotype | Clinical Significance |
Region | Description | Ref. | |||||
| Amino acid |
Nucleotide |
HCC (%) | Non-HCC (%) | P value | |||||||
| aa | mutations | nt | mutations | ||||||||
| AS | 4 | NC | 1383 | A1383C | B/C | HCC (postoperative survival in patients with HBV-HCC) (P = 0.028) | Independent predictors of HCC survival | [65] | |||
| 52.8 | 25.8 | 0.003 | miRNA binding site | [66] | |||||||
| AS | 5 | V5M/L | 1386 | G1386A/C | C | 50.0 | 4.9 | < 0.001 | [27] | ||
| 47.8 | 0.0 | < 0.001 | [38] | ||||||||
| 37.7 | 13.3 | 0.002 | [67] | ||||||||
| AS | 30 | NC | 1461 | G1461A/T/C | B/C | HCC (postoperative survival in patients with HBV-HCC) (P = 0.005) | B cell epitope | Independent predictors of HCC survival | [65] | ||
| AS | 36 | T36P/S/A | 1479 | A/G1479C/T/G | A/C/D | 49.3 | 22.7 | 0.034 | B cell epitope | B cell epitope | [66] |
| 15.3 | 7.8 | < 0.001 | B cell epitope | [66] | |||||||
| 80.0 | 17.1 | < 0.010 | [68] | ||||||||
| AS | 38 | P38S | 1485 | C1485T | B/C | 21.7 | 4.9 | 0.023 | B cell epitope | [27] | |
| HCC (postoperative survival in patients with HBV-HCC) (P = 0.006) | Independent predictors of HCC survival | [65] | |||||||||
| 48.7 | 13.9 | 0.001 | Independent risk factor for the HCC | [69] | |||||||
| 29.9 | 16.8 | 0.001 | B cell epitope | [66] | |||||||
| 30.4 | 15.0 | 0.038 | [67] | ||||||||
| AS | 44 | A44V | 1504, 1505 | C1504G, C1505G | A/D | 35.9 | 6.9 | 0.012 | B cell epitope | [70] | |
| AS | 50 | G50R | 1521 | G1521A/C | A/D | 60.0 | 4.3 | < 0.010 | [68] | ||
| AS | 57 | NC | 1544 | T1544A/C | B/C | HCC (postoperative survival in patients with HBV-HCC) (P = 0.039) | Independent predictors of HCC survival | [65] | |||
| AS | 80 | NC | 1613 | G1613A | B/C/C2 | HCC (postoperative survival in patients with HBV-HCC) (P = 0.006) | Core promoter | Independent predictors of HCC survival | [65] | ||
| 54.7 | 28.3 | 0.001 | Significance of association with HCC | [71] | |||||||
| 38.0 | 10.0 | < 0.050 | [72] | ||||||||
| AS | 81 | NC | 1613 | G1613A | B/C | 50.0 | 8.6 | 0.001 | Core promoter | [73] | |
| AS | 86 | NC | 1631 | C1631T | C | 8.3 | 1.8 | 0.010 | Core promoter | CP, NRE | [66] |
| AS | 94 | H94Y | 1653 | C1653T | B/C/C2 | 40.0 | 4.9 | < 0.001 | Box α, C/EBP,CCAAT/enhancing binding protein, Core promoter, EnhII | [27] | |
| HCC (postoperative survival in patients with HBV-HCC) (P = 0.015) | Independent predictors of HCC survival | [65] | |||||||||
| 61.3 | 25.3 | < 0.001 | Significance of association with HCC | [71] | |||||||
| 56.0 | 30.0 | 0.0013 | [74] | ||||||||
| 45.0 | 19.0 | < 0.050 | [72] | ||||||||
| 31.6 | 19.1 | 0.016 | Box α | [75] | |||||||
| 35.4 | 18.6 | < 0.001 | Box α, CP, C/EBP, EnII | [66] | |||||||
| 41.2 | 13.3 | < 0.001 | [67] | ||||||||
| 55.5 | 2.9 | < 0.001 | [73] | ||||||||
| 8.9 | 2.2 | 0.017 | [76] | ||||||||
| AS | 101 | S101Stop | 1675 | C1675A | B/C | 35.3 | 5.3 | 0.001 | Core promoter, EnhII | EnhII | [77] |
| AS | 106 | S106T | 1689 | T1689A | C2 | 19.3 | 4.4 | < 0.001 | Core promoter, EnhII | [76] | |
| AS | 116 | L/V116V/L | 1719 | T/G1719G/T | B/C | HCC (postoperative survival in patients with HBV-HCC) (P = 0.020) | BH3-like motif, Core promoter, EnhII, NRE | Independent predictors of HCC survival | [65] | ||
| 82.6 | 57.6 | 0.010 | BH3-like motif, CP, EnhII, HNF3, T cell epitope | [66] | |||||||
| AS | 117 | NC | 1724 | T1724C | B/C | 41.1 | 2.6 | 0.000 | BH3-like motif, Core promoter, EnhII, NRE | EnhII | [77] |
| AS | 118 | NC | 1727 | A1727G | D1 | 35.0 | 5.0 | 0.001 | BH3-like motif, Core promoter, EnhII, NRE | [78] | |
| AS | 123 | L123S | 1741 | T1741C | D1 | 30.0 | 7.5 | 0.006 | BH3-like motif, Core promoter, EnhII, NRE | [78] | |
| AS, DM | 127 | I127L/T/N/S | 1753 | T1753C/A | C | 36.7 | 12.2 | 0.007 | BH3-like motif, Core promoter, NRE | [27] | |
| 1753 | T1753C | B/C | HCC (postoperative survival in patients with HBV-HCC) (P = 0.047) | Independent predictors of HCC survival | [65] | ||||||
| 1752, 1753 | A1752C + T1753A/C/G | D | 52.2 | 20.0 | 0.033 | [79] | |||||
| 1753 | T1753A/C/G | C2 | 50.7 | 16.0 | < 0.001 | Significance of association with HCC | [71] | ||||
| 1753 | T1753A/C/G | C2 | 50.0 | 24.0 | 0.001 | [74] | |||||
| 1753 | T1753C/G | A/D | 43.6 | 17.2 | 0.041 | [70] | |||||
| 1753 | T1753A/C/G | B/C | 30.9 | 17.6 | 0.006 | EnhII/BCP | [75] | ||||
| 1753 | T1753A/C/G | C | 29.0 | 5.0 | < 0.001 | [67] | |||||
| AS | 130 | K130M | 1762 | A1762T | C2 | 94.7 | 74.7 | 0.001 | BH3-like motif, Core promoter | Significance of association with HCC | [71] |
| B/C | 80.0 | 25.7 | < 0.001 | [73] | |||||||
| AS | 131 | V131I | 1764 | G1764A | C2 | 98.7 | 78.7 | < 0.001 | BH3-like motif, Core promoter | Significance of association with HCC | [71] |
| B/C | 95.0 | 31.4 | < 0.001 | [73] | |||||||
| AS, DM | 134 | NC | 1773 | C1773T | D1 | 95.0 | 52.5 | 0.000 | Core promoter | [78] | |
| 1773, 1775 | C1773T + A1775G | D1 | 17.5 | 0.0 | 0.010 | [78] | |||||
| AS | 143 | C143R | 1800 | T1800C | C | 3.5 | 0.3 | 0.008 | Core promoter | CP | [66] |
| AS, DM | 100, 102 | NC | 1673, 1679 | C1673T + A1679G | D1 | 17.5 | 0.0 | 0.010 | Core promoter | [78] | |
| AS, TM | 128, 131 | NC + V131L | 1757, 1764, 1766 | G1757A, G1764C + C1766G | D1 | 37.5 | 12.5 | 0.010 | Core promoter | [78] | |
| AS, DM | 130, 131 | K130M + V131I | 1762, 1764 | A1762T + G1764A | C | 86.7 | 24.4 | < 0.001 | Core promoter | [27] | |
| D | HCC (HBV-DNA ≥ 5 log copies/mL) vs CLD (HBV-DNA < 5 log copies/mL) (P < 0.05) | [79] | |||||||||
| C2 | 91.0 | 73.0 | 0.0035 | [74] | |||||||
| A/D | 62.5 | 14.3 | 0.034 | [80] | |||||||
| A/D | 64.1 | 20.7 | 0.000 | [70] | |||||||
| B/C | 71.1 | 55.7 | 0.009 | EnhII/BCP | [75] | ||||||
| B/C | 64.0 | 50.8 | < 0.00001 | [81] | |||||||
| A/D | 44.9 | 20.7 | < 0.001 | [82] | |||||||
| C | 91.5 | 53.3 | < 0.001 | [67] | |||||||
| C2 | 60.7 | 22.2 | < 0.001 | [76] | |||||||
| Del, Ins | 129-154, 120-148, 115-149, 135-154, 137-151 | Deletion | 93-94 (4aa), 79-80 (2aa), 93-94 (4aa), 151-152 (3aa) | Insertion | C2 | HCC + LC (7.6%) vs CH + C (1.5%) (P = 0.017) | [30] | ||||
AA: Amino acid; AS: Amino acid substitution; BCP: Basal core promoter; C: Carrier; CH: Chronic hepatitis; CLD: Chronic liver disease; CP: Core protein; Del: Deletion; DM: Double mutation; EnhII: Enhancer II; HCC: Hepatocellular carcinoma; HNF3: Hepatocyte nuclear factor 3; Ins: Insertion; LC: Liver cirrhosis; miRNA: MicroRNA; NC: No change; NRE: Negative regulatory element; TM: Triple mutation.
HBx MUTATIONS RELATED TO CLINICAL SEVERITY
Mutations in EnhII and (or) the core promoter region (BCP mutation, T1753V, and C1653T)
In general, 3 types of mutations in the EnhII/BCP region [one mutation in EnhII (H94Y: C→T of nt 1653) and two mutations in BCP (I127L,T,N,S: T→V of nt 1753, K130M and V131I: A→T of nt 1762 and G→A of nt 1764)] are mutational “hot spots”, namely, the most frequently encountered among naturally occurring HBx mutations related to clinical severance from chronic hepatitis B patients, irrespective of genotype or geographical distributions (Table 1). The A1762T/G1764A BCP mutation leading to two overlapped HBx amino acid changes, K130M and V131I, is the most frequent HBV DNA mutation identified in many studies as being associated with HCC risk and outcomes[72,74,84-87]. The exact mechanism underlying the role of this mutation in hepatocarcinogenesis is still unknown. However, some underlying mechanisms have been recently elucidated. The mutation can cause a substantial decrease in HBeAg expression and enhancement of viral genome replication, which contribute to the liver disease progression via increased inflammation and viral invasion[88,89]. The mutation also leads to a truncated HBx protein, which not only promotes hepatocellular proliferation but also enhances HCC cell invasion and metastasis[90,91]. In particular, in chronic patients infected with genotype C2, this mutation is reported to be related to HBV genome deletion[26,31] or to be positively correlated with HBx M5V/L or H94Y mutations[28,37]. In addition, it may also contribute to hepatocarcinogenesis via reduced p21 expression, leading to rapid and uncontrolled cell proliferation[92]. A recent meta-analysis by Yang et al[82] revealed that the BCP mutation is present at significantly higher frequencies in HCC patients than in non-HCC controls, including patients with liver cirrhosis, chronic hepatitis and asymptomatic carriers. Our previous data using a Korean cohort with genotype C2-infected chronic patients also showed that the BCP mutation was the most frequently encountered mutation related to clinical severity (66.1%, 123/184 strains) and was significantly related to HCC [HCC (86.7%) vs chronic hepatitis (61%), P = 0.017; HCC (86.7%) vs asymptomatic carrier (24.4%), P < 0.001][28]. Our data also showed that during the natural course of HBV chronic infection, the most significant rise in the rate of the BCP mutation was found during the progression from asymptomatic carrier to chronic hepatitis (24.4%-61.0%), suggesting that the BCP mutation may play a major role in liver disease progression, especially in the progression from asymptomatic carrier to chronic hepatitis in chronic patients infected with genotype C2[28]. This finding has also been confirmed by a recent meta-analysis[82]. Yang et al[82] also demonstrated that HBV-infected patients with genotype C, including HCC patients, have a significantly higher risk of BCP mutation compared with those with genotype B, suggesting that the BCP mutation can increase the risk of HBV-related hepatocellular carcinoma, particularly in an HBV genotype C population.
An HBV genome transfection-based experiment indicated that the BCP mutation can reduce the synthesis of HBeAg and enhance viral replication. However, a meta-analysis and our previous report also showed that there is no significant difference in BCP mutation prevalence between HBeAg-positive and HBeAg-negative chronic HBV-infected patients[28,82], suggesting that BCP mutation may occur in the HBeAg-positive immune tolerance phase earlier than in the HBeAg-negative immune clearance phase, at least in chronic patients infected with genotype C2.
The other HCC-associated T1753V mutation (I127L,T,N,S: T→V of nt 1753) was also shown to affect HCC survival[93,94]. The mutations in the HBx protein, which include an I127L,T,N,S amino acid substitution, can change the HBx binding affinity to BCL2, thereby affecting HBx-induced apoptosis[95]. Our previous data using Korean HBV-infected patients with genotype C2 showed that the prevalence of this mutation was also significantly higher in chronic patients with severe liver disease, HCC or liver cirrhosis than in patients who had milder types of diseases, were carriers or had chronic hepatitis [HCC and LC (34.3%) vs chronic hepatitis and carrier (13.4%), P < 0.001][28]. The other study using chronic patients from India who had genotype A or D revealed that this mutation is also usually associated with advanced forms of liver disease and had an increased risk of HCC[69], suggesting that the T1753V mutation may play a significant role in liver disease progression. Our previous report showed that the T1753V mutation is significantly related to the BCP double mutation [patients with the BCP mutation (31.7%) vs patients without the BCP mutation (11.5%), P = 0.003], but not to HBeAg serostatus[28]. A recent multivariate survival analysis by Xie et al[66] showed that the T1753V mutation is an independent predictor of HCC survival.
The C1653T mutation, leading to a simultaneous H94Y amino acid change in HBx, is located in box α, which is a strong activation element of the EnhII/core promoter, can enhance the box α binding affinity and EnhII/core promoter activity[96,97]. Because many trans-regulated nuclear factors bind HBV at the 1653 site, this mutation can alter the binding affinity of these nuclear factors. The C1653T mutation has been recently reported to be a predictive factor for HCC in Japan[75,98] and is associated with fulminant hepatitis and the acute exacerbation of HCC[99,100]. A recent multivariate survival analysis by Xie et al[66] showed that the C1653T mutation together with the T1753V mutation is also an independent predictor of HCC survival. Furthermore, our previous report showed that the C1653T mutation is significantly related to advanced liver diseases in Korean patients with genotype C2 infections [patients with HCC or LC (36.3%) vs patients who have chronic hepatitis or are carriers (12.2%), P < 0.001]. It has been reported that the C1653T mutation, together with 1762T/1764A mutations, can reduce the pre-C mRNA level (to approximately 55%) and HBeAg secretion in a transient transfection system using Huh7 cells[101]. Our previous study also demonstrated that this mutation tended to be related to an HBeAg-negative serostatus (P = 0.087) and was significantly related to the BCP mutation [patients with the BCP mutation (35.0%) vs patients without the BCP mutation (6.6%), P < 0.001].
Mutation in the negative regulation domain of HBx (aa 1-50) (A1383C, G1386A/C-V5M/L, C1485T-P38S)
The A1383T synonymous mutation, which does not cause an amino acid change in the HBx protein, is located in the negative regulation domain of HBx (aa 1-50), and this mutation was first found to be associated with HCC in a Korean cohort[28]. In one clinical study using Chinese cohort mostly infected with genotype B and C, this mutation was also associated with a worse prognosis in patients after liver transplantation[66]. Recently, a comprehensive analysis study based on global data by Li et al[67] showed that A1383T is one of the HBx mutations identified as independent risk factors for genotype C HBV-related HCC. It has also been reported that tumor suppressor microRNA 15a/16 (miR-15a/16) can directly target the wildtype HBx RNA sequence (nt1362-1383), inducing Bcl-2 expression by acting as a sponge to bind and sequester endogenous miR-15a/16. Consequently, this mutation can lead to a reduced binding capacity of miR-15a/16 to the HBx protein[47], which can prevent the infected cell from apoptosis by altering critical cell signal pathways and thereby contributing to hepatocarcinogenesis.
The G1386A/C mutation leading to a simultaneous V5M/L amino acid change at codon 5 of the HBx protein was first introduced by our previous study using a Korean cohort with genotype C2 infections[28]. Our data showed that this mutation was significantly more frequently found in HCC patients than in patients in other disease groups. Notably, the prevalence of this mutation was abruptly increased in HCC patients rather than in liver cirrhosis patients during disease progression (HCC vs liver cirrhosis; 49.2% vs 25.6%, P = 0.024), strongly suggesting that this mutation is a genuine HCC-specific mutation that possibly plays a pivotal role in the progression from liver cirrhosis to HCC[28]. Recently, the combination of both BCP double mutations and both types of the V5M mutation, V5M and V5L, has also been reported to increase the HCC risk by 5.34 times compared with the wild type by inducing a higher NF-κB activity in transformed cells[86]. Our previous report showed that this mutation is significantly related to an HBeAg-negative serostatus [HBeAg-negative patients (40%) vs HBeAg-positive patients (19.1%), P = 0.004], suggesting that it may be generated from the immune clearance phase[28]. This mutation was also significantly related to the BCP mutation [patients with the BCP mutation (36.6%) vs patients without the BCP mutation (9.2%), P < 0.001]. To date, its clinical relevance has not been introduced except for a Korean cohort with genotype C2 infections. It is tempting to speculate that this mutation may play a pivotal role in hepatocarcinogenesis during the HBeAg-negative immune clearance phase during the natural course of genotype C2 HBV infection.
The C1485T mutation, leading to simultaneous P38S in the HBx protein, were first introduced as an independent risk factor for HCC development in a study by Muroyama et al[70] using a Japanese cohort with genotype C infections. Both studies using Korean cohorts with genotype C2 infections[28,68] and a recent investigation based on global data by Li et al[67] also revealed that this mutation is significantly related to HCC. A functional study supporting the relationship between the mutations with HCC still remains to be conducted. However, given that its mutation site is located at the B cell epitope region (Figure 1), this mutation may lead to persistent infection by providing a mechanism of evading the humoral immune response of the host.
Deletions or insertions in the C-terminal region of HBx
The C-terminal region of HBx plays a key role in controlling cell proliferation, viability, and transformation[102-105]. Therefore, C-terminally deleted or inserted HBx has reduced transactivation activity and inhibitory effects on cell proliferation and thus may contribute to HCC generation[106]. Moreover, its reduced transacting capacity might reduce HBV replication[107]. The C-terminal deletion or insertion is one of the most frequently reported mutations of HBx and has been frequently detected in tissues and serum samples from HCC patients, irrespective of genotype or geographical distribution[24,108,109]. Our previous report using a Korean cohort with genotype C2 infections showed that the prevalence of deletions or insertions was significantly higher in patients with severe liver disease, HCC, or cirrhosis of the liver (7.2%, 10/132), compared with patients who were carriers or had chronic hepatitis (1.5%, 2/135) (P = 0.017)[31]. All deletions in six strains were concentrated at the C-terminal end of HBx, encompassing the 113th to 154th codons. Four types of insertions (PKLL, GM, FFN, and tt) were observed in six patients. Notably, all insertions were accompanied by a BCP double mutation[31] (Figure 2). Furthermore, we first introduced a novel HBxAg vaccine escape mutation, X8Del with an 8-bp deletion in the C-terminal region of the HBx gene from 6 vaccinated Korean subjects[38]. Our in vitro and in vivo studies showed that this mutation causes a reduced secretion of HBsAg and HBV virions compared with the wild type, suggesting that the X8Del mutation may contribute to occult HBV infection in vaccinated individuals via the reduced secretion of HBsAg and virions, possibly by compromising the transacting capacity of HBxAg[38].
Figure 2.
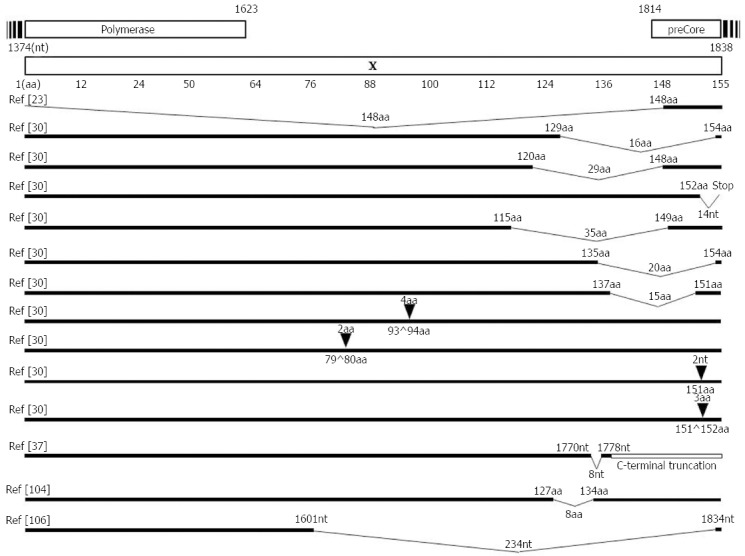
Mapping of deletions or insertions in the hepatitis B virus-X protein region. Deletions or insertions in the HBx region mainly occur in the C-terminal region of HBx, which may also contribute to the clinical outcome severity in chronic patients. HBx: Hepatitis B virus-X protein.
Other HBx mutations related to clinical severity
Recently, Xie et al[66] have reported 8 HBx mutational sites identified as significant independent risk predictors of HCC survival: 1383, 1461, 1485, 1544, 1613, 1653, 1719, and 1753 from a Chinese cohort mostly infected with genotype B and C. Despite the fact that the G1461V mutation is located at the B cell epitope, it (as a synonymous mutation) did not cause any simultaneous amino acid change in the HBx protein. Its regulatory modification in host cell or virion replication remains to be solved. The T1544V mutation also did not cause an amino acid change in the HBx protein. The G1613A mutation in the core promoter region is also a synonymous mutation, and its relationships with HCC have been reported in other previous studies.
Mutations in the BH-3-like motif of HBx can interfere with its interaction with two other Bcl-2 family members (Bcl-2 and Bcl-xL, which are critical for HBx to increase the intracellular calcium concentration), playing a significant role in viral replication and cell death[110]. Previous studies have reported that several types of mutations in the BH-3-like motif, T/G1719G/T, T1724C, and T1741C, were also significantly related to HCC[66,67,79].
The T1800C mutation leading to a simultaneous C143R amino acid change in the HBx protein is a novel genotype C HCC risk mutation identified by the Li et al[67] study, based on global data. To date, the function of this mutation in HCC remains unclear. However, of note, a recent study regarding HBV integration sites in 88 Chinese HCC patients showed that almost 40% of the integrated HBV genomes were cleaved at approximately nt1800, suggesting a potential role of this site in carcinogenesis, given that HBV genome integration has long been considered an important factor in HCC development.
CONCLUSION
In conclusion, HBx mutations may affect not only the HBx ORF but also the overlapped cis-elements. Considering all the HBx mutations related to clinical severity, the A1762T/G1764A BCP mutation is one of the most frequently encountered HBx mutations and plays a significant role in liver disease progression in chronic patients with HBV infections. It also further contributes to disease progression by inducing mutations of other HBx mutations related to clinical severity, such as G1386A/C (V5M/L), C1653T (H94Y), T1753V (I127V) and HBx C terminal deletion or insertion. Moreover, T1753V (I127L,T,N,S) and C1653T (H94Y) mutations in the EnhII/BCP region and A1383C, G1386A/C (V5M/L) and C1485T (P38S) in the negative regulation domain were significantly related to severe types of liver diseases, including HCC. Furthermore, deletions or insertions affecting the C-terminal region of HBx can also contribute to the clinical outcome severity in chronic patients. In addition, our recent study indicated that a novel mutation type (X8Del) composed of an 8-bp deletion in the C-terminal region of the HBx contributes to occult HBV infection in vaccinated Korean individuals via a reduced secretion of HBsAg and virions. Thus, several distinct types of HBx mutations may contribute to HBV pathogenesis by regulating HBV replication or host genes related to cell homeostasis.
Footnotes
Supported by National Research Foundation grant of Ministry of Science, ICT and Future Planning, South Korea, No. NRF-2015R1C1A1A02037267; and Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health and Welfare, South Korea, No. HI14C0955.
Conflict-of-interest statement: No conflict of interest.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: March 25, 2016
First decision: May 12, 2016
Article in press: June 2, 2016
P- Reviewer: Aghakhani A, Ma L, Wang K S- Editor: Ma YJ L- Editor: A E- Editor: Ma S
References
- 1.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 2.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guirgis BS, Abbas RO, Azzazy HM. Hepatitis B virus genotyping: current methods and clinical implications. Int J Infect Dis. 2010;14:e941–e953. doi: 10.1016/j.ijid.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 4.Gao S, Duan ZP, Coffin CS. Clinical relevance of hepatitis B virus variants. World J Hepatol. 2015;7:1086–1096. doi: 10.4254/wjh.v7.i8.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones SA, Hu J. Hepatitis B virus reverse transcriptase: diverse functions as classical and emerging targets for antiviral intervention. Emerg Microbes Infect. 2013;2:e56. doi: 10.1038/emi.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134:235–249. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 7.Datta S, Chatterjee S, Veer V, Chakravarty R. Molecular biology of the hepatitis B virus for clinicians. J Clin Exp Hepatol. 2012;2:353–365. doi: 10.1016/j.jceh.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moolla N, Kew M, Arbuthnot P. Regulatory elements of hepatitis B virus transcription. J Viral Hepat. 2002;9:323–331. doi: 10.1046/j.1365-2893.2002.00381.x. [DOI] [PubMed] [Google Scholar]
- 9.Park YM. Clinical utility of complex mutations in the core promoter and proximal precore regions of the hepatitis B virus genome. World J Hepatol. 2015;7:113–120. doi: 10.4254/wjh.v7.i1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quarleri J. Core promoter: a critical region where the hepatitis B virus makes decisions. World J Gastroenterol. 2014;20:425–435. doi: 10.3748/wjg.v20.i2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huan B, Siddiqui A. Regulation of hepatitis B virus gene expression. J Hepatol. 1993;17 Suppl 3:S20–S23. doi: 10.1016/s0168-8278(05)80419-2. [DOI] [PubMed] [Google Scholar]
- 12.Gupta N, Goyal M, Wu CH, Wu GY. The Molecular and Structural Basis of HBV-resistance to Nucleos(t)ide Analogs. J Clin Transl Hepatol. 2014;2:202–211. doi: 10.14218/JCTH.2014.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koumbi L. Current and future antiviral drug therapies of hepatitis B chronic infection. World J Hepatol. 2015;7:1030–1040. doi: 10.4254/wjh.v7.i8.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yano Y, Azuma T, Hayashi Y. Variations and mutations in the hepatitis B virus genome and their associations with clinical characteristics. World J Hepatol. 2015;7:583–592. doi: 10.4254/wjh.v7.i3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papatheodoridis GV, Chan HL, Hansen BE, Janssen HL, Lampertico P. Risk of hepatocellular carcinoma in chronic hepatitis B: assessment and modification with current antiviral therapy. J Hepatol. 2015;62:956–967. doi: 10.1016/j.jhep.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Glebe D, Geipel A. Selected phenotypic assays used to evaluate antiviral resistance and viral fitness of hepatitis B virus and its variants. Intervirology. 2014;57:225–231. doi: 10.1159/000360950. [DOI] [PubMed] [Google Scholar]
- 17.Luongo M, Critelli R, Grottola A, Gitto S, Bernabucci V, Bevini M, Vecchi C, Montagnani G, Villa E. Acute hepatitis B caused by a vaccine-escape HBV strain in vaccinated subject: sequence analysis and therapeutic strategy. J Clin Virol. 2015;62:89–91. doi: 10.1016/j.jcv.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 18.Locarnini SA, Yuen L. Molecular genesis of drug-resistant and vaccine-escape HBV mutants. Antivir Ther. 2010;15:451–461. doi: 10.3851/IMP1499. [DOI] [PubMed] [Google Scholar]
- 19.Delius H, Gough NM, Cameron CH, Murray K. Structure of the hepatitis B virus genome. J Virol. 1983;47:337–343. doi: 10.1128/jvi.47.2.337-343.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang TJ. Hepatitis B: the virus and disease. Hepatology. 2009;49:S13–S21. doi: 10.1002/hep.22881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidd-Ljunggren K, Miyakawa Y, Kidd AH. Genetic variability in hepatitis B viruses. J Gen Virol. 2002;83:1267–1280. doi: 10.1099/0022-1317-83-6-1267. [DOI] [PubMed] [Google Scholar]
- 22.Miyakawa Y, Mizokami M. Classifying hepatitis B virus genotypes. Intervirology. 2003;46:329–338. doi: 10.1159/000074988. [DOI] [PubMed] [Google Scholar]
- 23.Cho JH, Yoon KH, Lee KE, Park DS, Lee YJ, Moon HB, Lee KR, Choi CS, Cho EY, Kim HC. [Distribution of hepatitis B virus genotypes in Korea] Korean J Hepatol. 2009;15:140–147. doi: 10.3350/kjhep.2009.15.2.140. [DOI] [PubMed] [Google Scholar]
- 24.Kim H, Jee Y, Mun HS, Park JH, Yoon JH, Kim YJ, Lee HS, Hyun JW, Hwang ES, Cha CY, et al. Characterization of two hepatitis B virus populations in a single Korean hepatocellular carcinoma patient with an HBeAg-negative serostatus: a novel X-Gene-deleted strain with inverted duplication sequences of upstream enhancer site II. Intervirology. 2007;50:273–280. doi: 10.1159/000103915. [DOI] [PubMed] [Google Scholar]
- 25.Kim H, Jee Y, Mun HS, Song BC, Park JH, Hyun JW, Hwang ES, Cha CY, Kook YH, Kim BJ. Comparison of full genome sequences between two hepatitis B virus strains with or without preC mutation (A1896) from a single Korean hepatocellular carcinoma patient. J Microbiol Biotechnol. 2007;17:701–704. [PubMed] [Google Scholar]
- 26.Kim H, Jee YM, Song BC, Hyun JW, Mun HS, Kim HJ, Oh EJ, Yoon JH, Kim YJ, Lee HS, et al. Analysis of hepatitis B virus quasispecies distribution in a Korean chronic patient based on the full genome sequences. J Med Virol. 2007;79:212–219. doi: 10.1002/jmv.20789. [DOI] [PubMed] [Google Scholar]
- 27.Kim H, Jee YM, Song BC, Shin JW, Yang SH, Mun HS, Kim HJ, Oh EJ, Yoon JH, Kim YJ, et al. Molecular epidemiology of hepatitis B virus (HBV) genotypes and serotypes in patients with chronic HBV infection in Korea. Intervirology. 2007;50:52–57. doi: 10.1159/000096313. [DOI] [PubMed] [Google Scholar]
- 28.Kim HJ, Park JH, Jee Y, Lee SA, Kim H, Song BC, Yang S, Lee M, Yoon JH, Kim YJ, et al. Hepatitis B virus X mutations occurring naturally associated with clinical severity of liver disease among Korean patients with chronic genotype C infection. J Med Virol. 2008;80:1337–1343. doi: 10.1002/jmv.21219. [DOI] [PubMed] [Google Scholar]
- 29.Mun HS, Lee SA, Jee Y, Kim H, Park JH, Song BC, Yoon JH, Kim YJ, Lee HS, Hyun JW, et al. The prevalence of hepatitis B virus preS deletions occurring naturally in Korean patients infected chronically with genotype C. J Med Virol. 2008;80:1189–1194. doi: 10.1002/jmv.21208. [DOI] [PubMed] [Google Scholar]
- 30.Lee SA, Cho YK, Lee KH, Hwang ES, Kook YH, Kim BJ. Gender disparity in distribution of the major hydrophilic region variants of hepatitis B virus genotype C according to hepatitis B e antigen serostatus. J Med Virol. 2011;83:405–411. doi: 10.1002/jmv.21988. [DOI] [PubMed] [Google Scholar]
- 31.Lee SA, Mun HS, Kim H, Lee HK, Kim BJ, Hwang ES, Kook YH, Kim BJ. Naturally occurring hepatitis B virus X deletions and insertions among Korean chronic patients. J Med Virol. 2011;83:65–70. doi: 10.1002/jmv.21938. [DOI] [PubMed] [Google Scholar]
- 32.Mun HS, Lee SA, Kim H, Hwang ES, Kook YH, Kim BJ. Novel F141L pre-S2 mutation in hepatitis B virus increases the risk of hepatocellular carcinoma in patients with chronic genotype C infections. J Virol. 2011;85:123–132. doi: 10.1128/JVI.01524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim DW, Lee SA, Hwang ES, Kook YH, Kim BJ. Naturally occurring precore/core region mutations of hepatitis B virus genotype C related to hepatocellular carcinoma. PLoS One. 2012;7:e47372. doi: 10.1371/journal.pone.0047372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SA, Kim K, Kim H, Kim BJ. Nucleotide change of codon 182 in the surface gene of hepatitis B virus genotype C leading to truncated surface protein is associated with progression of liver diseases. J Hepatol. 2012;56:63–69. doi: 10.1016/j.jhep.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 35.Kim H, Lee SA, Kim DW, Lee SH, Kim BJ. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS One. 2013;8:e54486. doi: 10.1371/journal.pone.0054486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SA, Kim KJ, Kim DW, Kim BJ. Male-specific W4P/R mutation in the pre-S1 region of hepatitis B virus, increasing the risk of progression of liver diseases in chronic patients. J Clin Microbiol. 2013;51:3928–3936. doi: 10.1128/JCM.01505-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim BJ. Hepatitis B virus mutations related to liver disease progression of Korean patients. World J Gastroenterol. 2014;20:460–467. doi: 10.3748/wjg.v20.i2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim H, Gong JR, Lee SA, Kim BJ. Discovery of a Novel Mutation (X8Del) Resulting in an 8-bp Deletion in the Hepatitis B Virus X Gene Associated with Occult Infection in Korean Vaccinated Individuals. PLoS One. 2015;10:e0139551. doi: 10.1371/journal.pone.0139551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H, Hong SH, Lee SA, Gong JR, Kim BJ. Development of Fok-I based nested polymerase chain reaction-restriction fragment length polymorphism analysis for detection of hepatitis B virus X region V5M mutation. World J Gastroenterol. 2015;21:13360–13367. doi: 10.3748/wjg.v21.i47.13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim H, Kim BJ. Association of preS/S Mutations with Occult Hepatitis B Virus (HBV) Infection in South Korea: Transmission Potential of Distinct Occult HBV Variants. Int J Mol Sci. 2015;16:13595–13609. doi: 10.3390/ijms160613595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim H, Lee SA, Won YS, Lee H, Kim BJ. Occult infection related hepatitis B surface antigen variants showing lowered secretion capacity. World J Gastroenterol. 2015;21:1794–1803. doi: 10.3748/wjg.v21.i6.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H, Kim H, Lee SA, Won YS, Kim HI, Inn KS, Kim BJ. Upregulation of endoplasmic reticulum stress and reactive oxygen species by naturally occurring mutations in hepatitis B virus core antigen. J Gen Virol. 2015;96:1850–1854. doi: 10.1099/vir.0.000134. [DOI] [PubMed] [Google Scholar]
- 43.Lee SA, Kim H, Won YS, Seok SH, Na Y, Shin HB, Inn KS, Kim BJ. Male-specific hepatitis B virus large surface protein variant W4P potentiates tumorigenicity and induces gender disparity. Mol Cancer. 2015;14:23. doi: 10.1186/s12943-015-0303-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee SA, Kim KJ, Kim H, Choi WH, Won YS, Kim BJ. Hepatitis B virus preS1 deletion is related to viral replication increase and disease progression. World J Gastroenterol. 2015;21:5039–5048. doi: 10.3748/wjg.v21.i16.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu WH, Yeh SH, Chen PJ. Role of microRNAs in hepatitis B virus replication and pathogenesis. Biochim Biophys Acta. 2011;1809:678–685. doi: 10.1016/j.bbagrm.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K, Jacob ST. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu N, Zhang J, Jiao T, Li Z, Peng J, Cui Z, Ye X. Hepatitis B virus inhibits apoptosis of hepatoma cells by sponging the MicroRNA 15a/16 cluster. J Virol. 2013;87:13370–13378. doi: 10.1128/JVI.02130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu Z, Zhang Z, Doo E, Coux O, Goldberg AL, Liang TJ. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J Virol. 1999;73:7231–7240. doi: 10.1128/jvi.73.9.7231-7240.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, Protzer U, Hu Z, Jacob J, Liang TJ. Inhibition of cellular proteasome activities enhances hepadnavirus replication in an HBX-dependent manner. J Virol. 2004;78:4566–4572. doi: 10.1128/JVI.78.9.4566-4572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colgrove R, Simon G, Ganem D. Transcriptional activation of homologous and heterologous genes by the hepatitis B virus X gene product in cells permissive for viral replication. J Virol. 1989;63:4019–4026. doi: 10.1128/jvi.63.9.4019-4026.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci USA. 2010;107:4383–4388. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao Y, Da L, Tang H, Yang J, Lei Y, Tiollais P, Li T, Zhao M. Hepatitis B virus X protein reduces starvation-induced cell death through activation of autophagy and inhibition of mitochondrial apoptotic pathway. Biochem Biophys Res Commun. 2011;415:68–74. doi: 10.1016/j.bbrc.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 53.Li B, Gao B, Ye L, Han X, Wang W, Kong L, Fang X, Zeng Y, Zheng H, Li S, et al. Hepatitis B virus X protein (HBx) activates ATF6 and IRE1-XBP1 pathways of unfolded protein response. Virus Res. 2007;124:44–49. doi: 10.1016/j.virusres.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 54.Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- 55.McClain SL, Clippinger AJ, Lizzano R, Bouchard MJ. Hepatitis B virus replication is associated with an HBx-dependent mitochondrion-regulated increase in cytosolic calcium levels. J Virol. 2007;81:12061–12065. doi: 10.1128/JVI.00740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci USA. 2009;106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cougot D, Wu Y, Cairo S, Caramel J, Renard CA, Lévy L, Buendia MA, Neuveut C. The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J Biol Chem. 2007;282:4277–4287. doi: 10.1074/jbc.M606774200. [DOI] [PubMed] [Google Scholar]
- 58.Ren JH, Tao Y, Zhang ZZ, Chen WX, Cai XF, Chen K, Ko BC, Song CL, Ran LK, Li WY, et al. Sirtuin 1 regulates hepatitis B virus transcription and replication by targeting transcription factor AP-1. J Virol. 2014;88:2442–2451. doi: 10.1128/JVI.02861-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei C, Ni C, Song T, Liu Y, Yang X, Zheng Z, Jia Y, Yuan Y, Guan K, Xu Y, et al. The hepatitis B virus X protein disrupts innate immunity by downregulating mitochondrial antiviral signaling protein. J Immunol. 2010;185:1158–1168. doi: 10.4049/jimmunol.0903874. [DOI] [PubMed] [Google Scholar]
- 60.Jiang J, Tang H. Mechanism of inhibiting type I interferon induction by hepatitis B virus X protein. Protein Cell. 2010;1:1106–1117. doi: 10.1007/s13238-010-0141-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan J, Duan LX, Sun BS, Feitelson MA. Hepatitis B virus X protein protects against anti-Fas-mediated apoptosis in human liver cells by inducing NF-kappa B. J Gen Virol. 2001;82:171–182. doi: 10.1099/0022-1317-82-1-171. [DOI] [PubMed] [Google Scholar]
- 62.Kim JY, Song EH, Lee HJ, Oh YK, Choi KH, Yu DY, Park SI, Seong JK, Kim WH. HBx-induced hepatic steatosis and apoptosis are regulated by TNFR1- and NF-kappaB-dependent pathways. J Mol Biol. 2010;397:917–931. doi: 10.1016/j.jmb.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 63.Chirillo P, Pagano S, Natoli G, Puri PL, Burgio VL, Balsano C, Levrero M. The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc Natl Acad Sci USA. 1997;94:8162–8167. doi: 10.1073/pnas.94.15.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jung JK, Park SH, Jang KL. Hepatitis B virus X protein overcomes the growth-inhibitory potential of retinoic acid by downregulating retinoic acid receptor-beta2 expression via DNA methylation. J Gen Virol. 2010;91:493–500. doi: 10.1099/vir.0.015149-0. [DOI] [PubMed] [Google Scholar]
- 65.Arbuthnot P, Capovilla A, Kew M. Putative role of hepatitis B virus X protein in hepatocarcinogenesis: effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J Gastroenterol Hepatol. 2000;15:357–368. doi: 10.1046/j.1440-1746.2000.02069.x. [DOI] [PubMed] [Google Scholar]
- 66.Xie Y, Liu S, Zhao Y, Guo Z, Xu J. X protein mutations in hepatitis B virus DNA predict postoperative survival in hepatocellular carcinoma. Tumour Biol. 2014;35:10325–10331. doi: 10.1007/s13277-014-2331-0. [DOI] [PubMed] [Google Scholar]
- 67.Li W, Goto K, Matsubara Y, Ito S, Muroyama R, Li Q, Kato N. The characteristic changes in hepatitis B virus x region for hepatocellular carcinoma: a comprehensive analysis based on global data. PLoS One. 2015;10:e0125555. doi: 10.1371/journal.pone.0125555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cho EY, Choi CS, Cho JH, Kim HC. Association between Hepatitis B Virus X Gene Mutations and Clinical Status in Patients with Chronic Hepatitis B Infection. Gut Liver. 2011;5:70–76. doi: 10.5009/gnl.2011.5.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tuteja A, Siddiqui AB, Madan K, Goyal R, Shalimar V, Kaur N, Panda SK, Narayanasamy K, Subodh S, Acharya SK. Mutation profiling of the hepatitis B virus strains circulating in North Indian population. PLoS One. 2014;9:e91150. doi: 10.1371/journal.pone.0091150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muroyama R, Kato N, Yoshida H, Otsuka M, Moriyama M, Wang Y, Shao RX, Dharel N, Tanaka Y, Ohta M, et al. Nucleotide change of codon 38 in the X gene of hepatitis B virus genotype C is associated with an increased risk of hepatocellular carcinoma. J Hepatol. 2006;45:805–812. doi: 10.1016/j.jhep.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 71.Asim M, Malik A, Sarma MP, Polipalli SK, Begum N, Ahmad I, Khan LA, Husain SA, Akhtar N, Husain S, et al. Hepatitis B virus BCP, Precore/core, X gene mutations/genotypes and the risk of hepatocellular carcinoma in India. J Med Virol. 2010;82:1115–1125. doi: 10.1002/jmv.21774. [DOI] [PubMed] [Google Scholar]
- 72.Jang JW, Chun JY, Park YM, Shin SK, Yoo W, Kim SO, Hong SP. Mutational complex genotype of the hepatitis B virus X /precore regions as a novel predictive marker for hepatocellular carcinoma. Cancer Sci. 2012;103:296–304. doi: 10.1111/j.1349-7006.2011.02170.x. [DOI] [PubMed] [Google Scholar]
- 73.Tatsukawa M, Takaki A, Shiraha H, Koike K, Iwasaki Y, Kobashi H, Fujioka S, Sakaguchi K, Yamamoto K. Hepatitis B virus core promoter mutations G1613A and C1653T are significantly associated with hepatocellular carcinoma in genotype C HBV-infected patients. BMC Cancer. 2011;11:458. doi: 10.1186/1471-2407-11-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu Y, Jin Y, Guo X, Bai X, Chen T, Wang J, Qian G, Groopman JD, Gu J, Li J, et al. Comparison study on the complete sequence of hepatitis B virus identifies new mutations in core gene associated with hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2010;19:2623–2630. doi: 10.1158/1055-9965.EPI-10-0469. [DOI] [PubMed] [Google Scholar]
- 75.Shinkai N, Tanaka Y, Ito K, Mukaide M, Hasegawa I, Asahina Y, Izumi N, Yatsuhashi H, Orito E, Joh T, et al. Influence of hepatitis B virus X and core promoter mutations on hepatocellular carcinoma among patients infected with subgenotype C2. J Clin Microbiol. 2007;45:3191–3197. doi: 10.1128/JCM.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qu LS, Zhu J, Liu TT, Shen XZ, Chen TY, Ni ZP, Ni RZ, Lu CH. Effect of combined mutations in the enhancer II and basal core promoter of hepatitis B virus on development of hepatocellular carcinoma in Qidong, China. Hepatol Res. 2014;44:1186–1195. doi: 10.1111/hepr.12291. [DOI] [PubMed] [Google Scholar]
- 77.Kim JK, Chang HY, Lee JM, Baatarkhuu O, Yoon YJ, Park JY, Kim DY, Han KH, Chon CY, Ahn SH. Specific mutations in the enhancer II/core promoter/precore regions of hepatitis B virus subgenotype C2 in Korean patients with hepatocellular carcinoma. J Med Virol. 2009;81:1002–1008. doi: 10.1002/jmv.21501. [DOI] [PubMed] [Google Scholar]
- 78.Fan W, Shi B, Wei H, Du G, Song S. Comparison of hepatitis B X gene mutation between patients with hepatocellular carcinoma and patients with chronic hepatitis B. Virus Genes. 2011;42:162–170. doi: 10.1007/s11262-010-0557-5. [DOI] [PubMed] [Google Scholar]
- 79.Khan A, Al Balwi MA, Tanaka Y, Hajeer A, Sanai FM, Al Abdulkarim I, Al Ayyar L, Badri M, Saudi D, Tamimi W, et al. Novel point mutations and mutational complexes in the enhancer II, core promoter and precore regions of hepatitis B virus genotype D1 associated with hepatocellular carcinoma in Saudi Arabia. Int J Cancer. 2013;133:2864–2871. doi: 10.1002/ijc.28307. [DOI] [PubMed] [Google Scholar]
- 80.Elkady A, Tanaka Y, Kurbanov F, Oynsuren T, Mizokami M. Virological and clinical implication of core promoter C1752/V1753 and T1764/G1766 mutations in hepatitis B virus genotype D infection in Mongolia. J Gastroenterol Hepatol. 2008;23:474–481. doi: 10.1111/j.1440-1746.2008.05321.x. [DOI] [PubMed] [Google Scholar]
- 81.Venard V, Corsaro D, Kajzer C, Bronowicki JP, Le Faou A. Hepatitis B virus X gene variability in French-born patients with chronic hepatitis and hepatocellular carcinoma. J Med Virol. 2000;62:177–184. doi: 10.1002/1096-9071(200010)62:2<177::aid-jmv8>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 82.Yang Z, Zhuang L, Lu Y, Xu Q, Tang B, Chen X. Naturally occurring basal core promoter A1762T/G1764A dual mutations increase the risk of HBV-related hepatocellular carcinoma: a meta-analysis. Oncotarget. 2016;7:12525–12536. doi: 10.18632/oncotarget.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Malik A, Singhal DK, Albanyan A, Husain SA, Kar P. Hepatitis B virus gene mutations in liver diseases: a report from New Delhi. PLoS One. 2012;7:e39028. doi: 10.1371/journal.pone.0039028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen QY, Harrison TJ, Sabin CA, Li GJ, Huang GM, Yang JY, Wang XY, Li H, Liu MH, Fang ZL. The Effect of HBV Genotype C on the Development of HCC Differs Between Wild-Type Viruses and Those With BCP Double Mutations (T(1762)A(1764)) Hepat Mon. 2014;14:e16214. doi: 10.5812/hepatmon.16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Muñoz A, Chen JG, Egner PA, Marshall ML, Johnson JL, Schneider MF, Lu JH, Zhu YR, Wang JB, Chen TY, et al. Predictive power of hepatitis B 1762T/1764A mutations in plasma for hepatocellular carcinoma risk in Qidong, China. Carcinogenesis. 2011;32:860–865. doi: 10.1093/carcin/bgr055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee JH, Han KH, Lee JM, Park JH, Kim HS. Impact of hepatitis B virus (HBV) x gene mutations on hepatocellular carcinoma development in chronic HBV infection. Clin Vaccine Immunol. 2011;18:914–921. doi: 10.1128/CVI.00474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Datta S, Banerjee A, Chandra PK, Biswas A, Panigrahi R, Mahapatra PK, Panda CK, Chakrabarti S, Bhattacharya SK, Chakravarty R. Analysis of hepatitis B virus X gene phylogeny, genetic variability and its impact on pathogenesis: implications in Eastern Indian HBV carriers. Virology. 2008;382:190–198. doi: 10.1016/j.virol.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 88.Buckwold VE, Xu Z, Chen M, Yen TS, Ou JH. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J Virol. 1996;70:5845–5851. doi: 10.1128/jvi.70.9.5845-5851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hakami A, Ali A, Hakami A. Effects of hepatitis B virus mutations on its replication and liver disease severity. Open Virol J. 2013;7:12–18. doi: 10.2174/1874357901307010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yeh CT, So M, Ng J, Yang HW, Chang ML, Lai MW, Chen TC, Lin CY, Yeh TS, Lee WC. Hepatitis B virus-DNA level and basal core promoter A1762T/G1764A mutation in liver tissue independently predict postoperative survival in hepatocellular carcinoma. Hepatology. 2010;52:1922–1933. doi: 10.1002/hep.23898. [DOI] [PubMed] [Google Scholar]
- 91.Chen L, Zhang Q, Chang W, Du Y, Zhang H, Cao G. Viral and host inflammation-related factors that can predict the prognosis of hepatocellular carcinoma. Eur J Cancer. 2012;48:1977–1987. doi: 10.1016/j.ejca.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 92.Kwun HJ, Jang KL. Natural variants of hepatitis B virus X protein have differential effects on the expression of cyclin-dependent kinase inhibitor p21 gene. Nucleic Acids Res. 2004;32:2202–2213. doi: 10.1093/nar/gkh553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu S, Zhang H, Gu C, Yin J, He Y, Xie J, Cao G. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101:1066–1082. doi: 10.1093/jnci/djp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu L, Qian G, Tang L, Su J, Wang JS. Genetic variations of hepatitis B virus and serum aflatoxin-lysine adduct on high risk of hepatocellular carcinoma in Southern Guangxi, China. J Hepatol. 2010;53:671–676. doi: 10.1016/j.jhep.2010.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Geng X, Harry BL, Zhou Q, Skeen-Gaar RR, Ge X, Lee ES, Mitani S, Xue D. Hepatitis B virus X protein targets the Bcl-2 protein CED-9 to induce intracellular Ca2+ increase and cell death in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2012;109:18465–18470. doi: 10.1073/pnas.1204652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.López-Cabrera M, Letovsky J, Hu KQ, Siddiqui A. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1990;87:5069–5073. doi: 10.1073/pnas.87.13.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yuh CH, Chang YL, Ting LP. Transcriptional regulation of precore and pregenomic RNAs of hepatitis B virus. J Virol. 1992;66:4073–4084. doi: 10.1128/jvi.66.7.4073-4084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tanaka Y, Mukaide M, Orito E, Yuen MF, Ito K, Kurbanov F, Sugauchi F, Asahina Y, Izumi N, Kato M, et al. Specific mutations in enhancer II/core promoter of hepatitis B virus subgenotypes C1/C2 increase the risk of hepatocellular carcinoma. J Hepatol. 2006;45:646–653. doi: 10.1016/j.jhep.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 99.Kaneko M, Uchida T, Moriyama M, Arakawa Y, Shikata T, Gotoh K, Mima S. Probable implication of mutations of the X open reading frame in the onset of fulminant hepatitis B. J Med Virol. 1995;47:204–208. doi: 10.1002/jmv.1890470304. [DOI] [PubMed] [Google Scholar]
- 100.Uchida T, Saitoh T, Shinzawa H. Mutations of the X region of hepatitis B virus and their clinical implications. Pathol Int. 1997;47:183–193. doi: 10.1111/j.1440-1827.1997.tb04479.x. [DOI] [PubMed] [Google Scholar]
- 101.Günther S, Piwon N, Will H. Wild-type levels of pregenomic RNA and replication but reduced pre-C RNA and e-antigen synthesis of hepatitis B virus with C(1653) --> T, A(1762) --> T and G(1764) --> A mutations in the core promoter. J Gen Virol. 1998;79(Pt 2):375–380. doi: 10.1099/0022-1317-79-2-375. [DOI] [PubMed] [Google Scholar]
- 102.Bréchot C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology. 2004;127:S56–S61. doi: 10.1053/j.gastro.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 103.Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene. 2006;25:3823–3833. doi: 10.1038/sj.onc.1209559. [DOI] [PubMed] [Google Scholar]
- 104.Feitelson MA, Bonamassa B, Arzumanyan A. The roles of hepatitis B virus-encoded X protein in virus replication and the pathogenesis of chronic liver disease. Expert Opin Ther Targets. 2014;18:293–306. doi: 10.1517/14728222.2014.867947. [DOI] [PubMed] [Google Scholar]
- 105.Zhang H, Shan CL, Li N, Zhang X, Zhang XZ, Xu FQ, Zhang S, Qiu LY, Ye LH, Zhang XD. Identification of a natural mutant of HBV X protein truncated 27 amino acids at the COOH terminal and its effect on liver cell proliferation. Acta Pharmacol Sin. 2008;29:473–480. doi: 10.1111/j.1745-7254.2008.00764.x. [DOI] [PubMed] [Google Scholar]
- 106.Ali A, Abdel-Hafiz H, Suhail M, Al-Mars A, Zakaria MK, Fatima K, Ahmad S, Azhar E, Chaudhary A, Qadri I. Hepatitis B virus, HBx mutants and their role in hepatocellular carcinoma. World J Gastroenterol. 2014;20:10238–10248. doi: 10.3748/wjg.v20.i30.10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou F, Xu H, Chen M, Xiao H, Zhang Z, Lu Y, Ren J, Dong J. X gene/core promoter deletion mutation: a novel mechanism leading to hepatitis B ‘e’ antigennegative chronic hepatitis B. Mol Med Rep. 2014;10:799–803. doi: 10.3892/mmr.2014.2248. [DOI] [PubMed] [Google Scholar]
- 108.Wang WL, London WT, Feitelson MA. Hepatitis B x antigen in hepatitis B virus carrier patients with liver cancer. Cancer Res. 1991;51:4971–4977. [PubMed] [Google Scholar]
- 109.Chen GG, Li MY, Ho RL, Chak EC, Lau WY, Lai PB. Identification of hepatitis B virus X gene mutation in Hong Kong patients with hepatocellular carcinoma. J Clin Virol. 2005;34:7–12. doi: 10.1016/j.jcv.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 110.Geng X, Huang C, Qin Y, McCombs JE, Yuan Q, Harry BL, Palmer AE, Xia NS, Xue D. Hepatitis B virus X protein targets Bcl-2 proteins to increase intracellular calcium, required for virus replication and cell death induction. Proc Natl Acad Sci USA. 2012;109:18471–18476. doi: 10.1073/pnas.1204668109. [DOI] [PMC free article] [PubMed] [Google Scholar]


