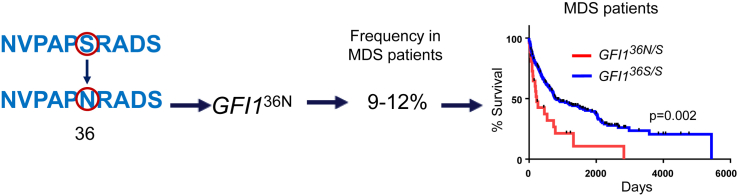
Abstract
Inherited gene variants play an important role in malignant diseases. The transcriptional repressor growth factor independence 1 (GFI1) regulates hematopoietic stem cell (HSC) self-renewal and differentiation. A single-nucleotide polymorphism of GFI1 (rs34631763) generates a protein with an asparagine (N) instead of a serine (S) at position 36 (GFI136N) and has a prevalence of 3%–5% among Caucasians. Because GFI1 regulates myeloid development, we examined the role of GFI136N on the course of MDS disease. To this end, we determined allele frequencies of GFI136N in four independent MDS cohorts from the Netherlands and Belgium, Germany, the ICGC consortium, and the United States. The GFI136N allele frequency in the 723 MDS patients genotyped ranged between 9% and 12%. GFI136N was an independent adverse prognostic factor for overall survival, acute myeloid leukemia-free survival, and event-free survival in a univariate analysis. After adjustment for age, bone marrow blast percentage, IPSS score, mutational status, and cytogenetic findings, GFI136N remained an independent adverse prognostic marker. GFI136S homozygous patients exhibited a sustained response to treatment with hypomethylating agents, whereas GFI136N patients had a poor sustained response to this therapy. Because allele status of GFI136N is readily determined using basic molecular techniques, we propose inclusion of GFI136N status in future prospective studies for MDS patients to better predict prognosis and guide therapeutic decisions.
Graphical abstract
Highlights
-
•
GFI136N is present in about 9%–12% of all Caucasian patients with myelodysplastic syndrome.
-
•
GFI136N is an independent, adverse prognostic factor for survival.
-
•
GFI136N patients with myelodysplastic syndrome respond poorly to hypomethylating agents.
GFI1 is a zinc finger transcriptional repressor that recruits histone-modifying enzymes, such as histone deacetylases, to the loci of its target genes 1, 2. GFI1 regulates the functions of hematopoietic stem cells (HSCs) 1, 3 as well as myeloid–lymphoid lineage decisions 4, 5. A variant form of the GFI1 gene (denominated GFI136N) is associated with a predisposition to develop de novo acute myeloid leukemia (AML) [6], but it has also been reported to be involved in a case of neutropenia [7]. Taking into consideration the predisposing role of GFI136N to de novo AML and its role in myeloid development, we investigated the role of GFI136N in myelodysplastic syndrome (MDS).
Methods
Patient cohort
All patient samples (peripheral blood [PB] or bone marrow [BM] aspirates) were obtained with informed consent according to the Declaration of Helsinki. The respective local ethics committees approved the use of all patient samples.
The clinical characteristics of patients with a confirmed diagnosis of MDS used in this study have been previously described 8, 9, 10, 11, 12, 13. Events in “event-free survival” were defined as death from any cause or progression of MDS to AML with blast counts higher than 20%. Overall survival events are defined as death from any cause.
Bone marrow morphology and cytopenia classification
Bone marrow morphology studies were performed at individual centers. MDS was classified based on the World Health Organization (WHO) definition [14].
Genotyping
Genotyping was performed according to published procedures [6].
Statistical methods
Significance of differences in percentages was determined using the two-sided, two-sample t test. Survival of the different human cohorts is based on the presence of GFI136N univariate analysis using Kaplan–Meier survival methods. Differences were assessed using the log-rank (Mantel–Cox) test. We used Cox proportional-hazards regression modeling to determine the influence of different factors with respect to survival. Factors taken into account were International Prognostic Scoring System (IPSS) risk group, BM blast count, age, sex, cytogenetic findings (based on IPSS classification), and in a last step, presence of GFI136N. Analyses were performed either separately (with each factor analyzed independently) or with the presence or absence of GFI136N. All p values reported are two-sided. Because of the exploratory nature of this study, no adjustment for multiple testing was done. All analyses presented were performed using GraphPad Prism 6 software (GraphPad Software, La Jolla, CA) or SPSS Version 19 (IBM, Armonk, NY).
Results
Between 9% and 12% of all adult MDS patients in four different Caucasian cohorts from Europe and the United States 8, 9, 10, 11, 12 were heterozygous for GFI136N, and only two patients from the European cohorts (and none of the U.S. cohort) were homozygous for GFI136N (Table 1). GFI136N allele frequency was higher among MDS patients than among the control cohort (3%–5%) as reported in our previous study regarding the role of GFI136N in de novo AML [15]. Although we did not determine the frequency of GFI136N among sex- and age-matched control subjects from every region, it is possible that GFI136N predisposes to MDS, similar to its predisposing role in de novo AML [15].
Table 1.
Features of GFI136N- and GFI136S-carrying adult MDS patientsa
| GFI136N (homo [2 patients]- or heterozygous) | GFI136S (homozygozus) | P value | |
|---|---|---|---|
| % All countries | 10 (n = 75) | 90 (n = 648) | |
| % United States | 11 (n = 30) | 89 (n = 254) | |
| % Germany | 9 (n = 20) | 91 (n = 193) | |
| % Netherlands and Belgium | 12 (n = 11) | 88 (n = 84) | |
| % ICGC | 11 (n = 14) | 89 (n = 117) | |
| Mean age | 66 ± 1.5 (n = 73) | 62 ± 0.6 (n = 638) | 0.01 |
| Gender (% male) | 64 (n = 47) | 66 (n = 426) | 0.7 |
| Blast percentage (BM) WHO | 9.4 ± 0.3 (n = 48) | 6.7 ± 1.1 (n = 464) | 0.01 |
| Hemoglobin (mg/dL) | 8.4 ± 0.5 (n = 24) | 8.1 ± 0.1 (n = 191) | 0.57 |
| Platelet count (1/nL) | 186 ± 37 (n = 24) | 177 ± 11 (n = 195) | 0.8 |
| Neutrophil count (1/nL) | 2.8 ± 0.8 (n = 24) | 2.7 ± 0.2 (n = 196) | 0.9 |
| Cytogenetic low risk (%) | 46 (n = 27) | 62 (n = 246) | 0.03 |
| Cytogenetic intermediate risk (%) | 14 (n = 8) | 16 (n = 64) | 0.5 |
| Cytogenetic high risk (%) | 41 (n = 24) | 24 (n = 96) | 0.006 |
| IPSS low (%) | 25 (n = 13) | 25 (n = 127) | 0.7 |
| IPSS intermediate 1 (%) | 40 (n = 38) | 42 (n = 323) | 0.7 |
| IPSS intermediate 2 (%) | 21 (n = 11) | 21 (n = 109) | 0.7 |
| IPSS high (%) | 15 (n = 8) | 10 (n = 52) | 0.25 |
| 5q– (%) | 5 (n = 2) | 4 (n = 13) | 0.8 |
| RA (%) | 0 (n = 0) | 9 (n = 30) | 0.054 |
| RARS+ RARST (%) | 11 (n = 4) | 9 (n = 28) | 0.7 |
| RAEB-1 (%) | 22 (n = 8) | 18 (n = 59) | 0.65 |
| RAEB-2 (%) | 39 (n = 15) | 22 (n = 69) | 0.02 |
| RAEB-1 + RAEB-2 (%) | 61 (23) | 40 (128) | 0.01 |
| RCMD (%) | 24 (n = 9) | 38 (n = 123) | 0.09 |
| MDS-u (%) | 0 | 1 (n = 4) | 0.5 |
BM = bone marrow; ICGC = International Cancer Genome Consortium; WHO = World Health Organization.
GFI136N includes patients who are either homozygous or heterozygous for GFI136N and, thus, carrying one GFI136S allele. GFI136S refers to patients homozygous for GFI136S. Cytogenetic low risk: normal karyotype, 5q–, 20q–, –Y; poor risk: complex aberrations (≥3 anomalies), chromosome 7 anomalies; intermediate risk: all other aberrations. IPSS was based on Greenberg et al. [17]. Refractory anemia (RA), refractory anemia with ring sideroblasts (RARS), refractory anemia with excess blasts, type 1 (RAEB-1), refractory anemia with excess blasts, type 2 (RAEB-2), refractory cytopenia with multilineage dysplasia (RCMD), and MDS-unclassified (MDS-u) are based on the WHO definition for MDS. Standard errors of mean are given. Data for IPPS and cytogenetic classification are missing due to the lack of cytogenetic information on patients at the time of diagnosis. Data for histologic classification are missing because of the missing specification of MDS according to WHO criteria. The missing patients were diagnosed as having MDS according to the WHO definition. Student's t test was used to determine the significance of values, and two-sample t tests were used to determine the significance of differences between percentages. The different cohorts were independent of each other. No overlap exists with respect to samples. The numbers in parentheses correspond to the absolute numbers related to the indicated percentages.
We analyzed the effect of GFI136N on MDS disease course in two independent cohorts. Patients were recruited and treated either in the United States or in Europe. The European cohorts consisted of patients recruited and treated at different centers in Germany, Belgium, and The Netherlands 8, 9, 10, 11, 12. The U.S. cohorts were referred to the Cleveland Clinic.
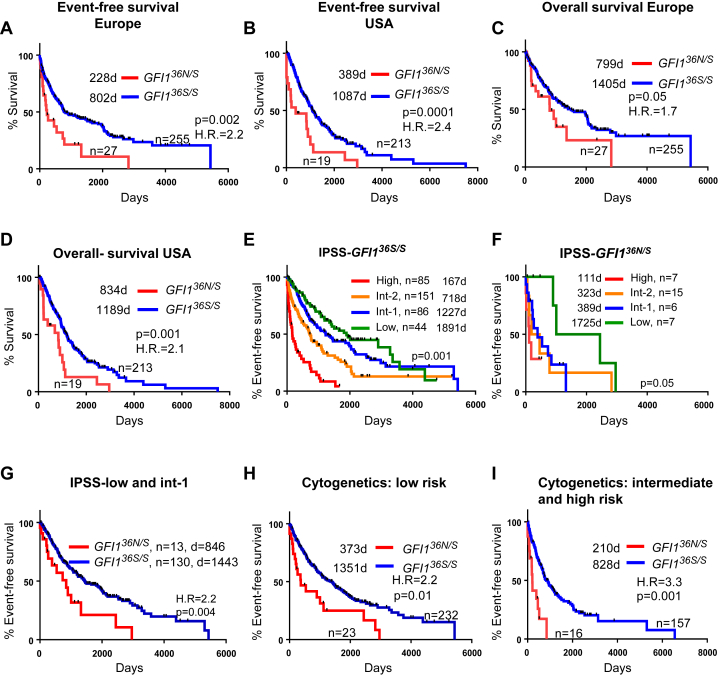
In the two MDS cohorts from Europe and the United States, presence of GFI136N was associated with an inferior event-free survival rate (Fig. 1A, B). GFI136N also had a negative impact on overall survival (Fig. 1C, D) and on AML-free survival (data not shown).
Figure 1.
Correlation between GFI136N and disease course of patients with MDS. (A) Patients from different European cohorts diagnosed with MDS on the basis of WHO criteria were genotyped with respect to the presence of GFI136N and examined with respect to median event-free survival (see also Methods); 95% confidence interval (CI) = 1.6–5.6. Median survival is indicated. (B) MDS Patients from a U.S. cohort diagnosed with MDS on the basis of WHO criteria were genotyped with respect to the presence of GFI136N and examined with respect to median event-free survival; 95% CI = 1.7–7.2. Median survival is indicated. (C) The same cohorts as in (A) were examined with respect to overall survival (death of any cause); 95% CI = 1.0–3.9. Median survival is indicated. (D) The same cohort as in (B) was examined with respect to overall survival (death from any cause) 95% CI = 1.5–5.8. Median survival is indicated. (E) Event-free survival of GFI136S homozygous patients was stratified based on IPSS classification. No sufficient follow-up was available for the International Cancer Genome Consortium (ICGC) patients. Follow-up is based on the patient cohorts from the United States, the Netherlands, Belgium, and Germany. Median survival is indicated. (F) Event-free survival of GFI136N homozygous or heterozygous patients was stratified based on IPSS classification. No sufficient follow up was available for the ICGC patients. Follow-up is based on the patient cohorts from the United States, the Netherlands, Belgium, and Germany. Median survival is indicated. (G) Event-free survival of patients (shown in A) classified as either IPSS subtype low or intermediate 1 (int-1) was stratified with respect to the presence of GFI136N; 95% CI = 1.4–6.2. (H) Patients from the U.S. and European cohorts with cytogenetic risk characteristics belonging to subtype “low” were stratified by the presence of GFI136N with respect to event-free survival; 95% CI = 1.5–5.7. Median survival is indicated. (I) Patients from the U.S. and European cohorts with cytogenetic risk characteristics belonging to subtype “intermediate” or “high” were stratified by presence of GFI136N with respect to event-free survival; 95% CI = 3.3–23.3. Median survival is indicated.
Next, we examined the association between GFI136N and established prognostic factors. GFI136N carriers were older, exhibited a tendency toward higher BM blast counts at diagnosis, were diagnosed with a more advanced stage of the disease according to histologic parameters, and had more adverse cytogenetic findings (Table 1). With respect to key blood parameters, no other differences between GFI136N and GFI136S carriers were observed (Table 1).
American and European GFI136S homozygous patients had median follow-ups of 1,100 and 975 days, respectively. American and European GFI136N carriers had median follow-ups of 540 and 350 days, respectively. To gain more statistical power and to perform more specific analysis, we combined the U.S. and European cohorts. One approach used to predict outcome of MDS patients is based on IPSS 16, 17. A recently introduced, revised version of IPSS (denominated IPSS-R) distinguishes between more subclasses based on cytogenetic findings and cytopenic lineages [16]. However, not all of the specific data for determining IPSS-R status were present in our databases. Therefore, we focused our examination on the nonrevised version of IPSS.
As reported previously [17], IPSS scoring predicted event-free outcome of GFI136S homozygous patients (Fig. 1E). On the basis of the same scoring system, GFI136N carriers had a significantly shorter event-free survival (Fig. 1F). Especially among MDS patients in the low-risk groups (groups low and intermediate 1 based on IPSS), GFI136N carriers had a much shorter event-free (Fig. 1G) and AML-free survival (data not shown) than GFI136S homozygous patients. We also examined the association between allele status, cytogenetic findings and event-free survival. Similarly, presence of GFI136N was associated with an inferior outcome, independent of the cytogenetic finding (Table 1). After stratification for cytogenetic risk groups (“low” as one group and “intermediate and high” as a second group), the presence of a GFI136N allele was again linked to inferior event-free survival (Fig. 1H, I) in both comparisons.
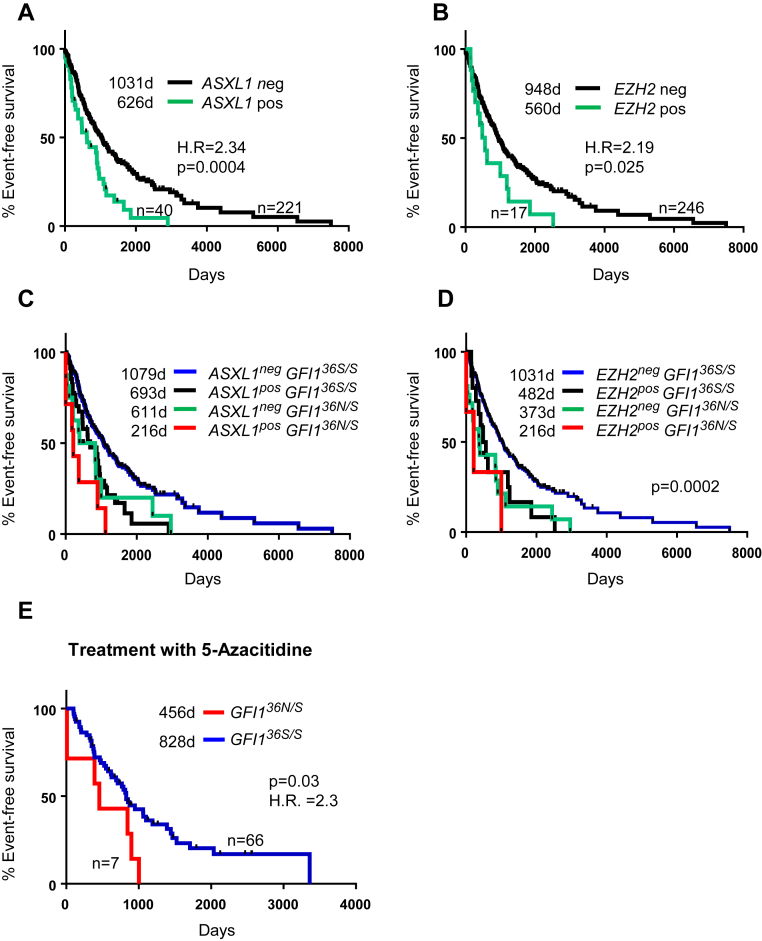
MDS patients with somatic mutations within EZH2 or ASXL1 have an inferior prognosis 18, 19 (Fig. 2A, B). Presence of GFI136N in the absence of ASXL1 or EZH2 mutations had a similar effect on event-free survival as the presence of ASXL1 or EZH2 mutations (Fig. 2C, D). Furthermore, the combined presence of a mutated form of either ASXL1 or EZH2 and a GFI136N allele had an additional adverse effect on event-free survival (Fig. 2C, D). Similar analyses were not possible for mutations of P53 or RUNX1 18, 19, because of the small number of cases with P53 or RUNX1 mutation and GFI136N.
Figure 2.
Association between GFI136N, mutational status, prognosis, and therapeutic response. (A) Median event-free survival of patients based on mutational status of ASXL1; 95% confidence interval (CI) = 1.5–5.7. (B) Median event-free survival of patients based on mutational status of EZH2; 95% CI = 3.4–23.3. (C) Median event-free survival of patients based on presence of GFI136N and mutational status of ASXL1. (D) Median event-free survival of patients based on presence of GFI136N and mutational status of EZH2. (E) From the cohorts of patients treated in Europe and the United States, patients treated with 5-azacitidine were stratified with respect to GFI1 status. Presence of GFI136N was associated with a shorter response. Median survival is indicated; 95% CI = 1.1–10.5.
Because MDS patients who are hetero- or homozygous for GFI136N tend to be older at diagnosis, have more frequent adverse cytogenetic findings, and have a higher blast cell count at diagnosis, we examined whether the presence of GFI136N represents an adverse prognostic factor after adjusting for these findings. We found that GFI136N was an independent marker after adjusting for the variables IPSS score, cytogenetic findings, and age, either alone or in combination (Supplementary Table E1, Supplementary Table E2, online only, available at www.exphem.org).
Discussion
One possible explanation why GFI136N accelerates AML development in MDS patients could be based on our findings using murine models. We previously generated mice expressing GFI136N or GFI136S instead of murine Gfi1 [15]. We reported that GFI136N is not able to bind to its target genes to the same extent as the more common “wild-type” GFI136S variant. Hence, presence of one allele of GFI136N led to higher genomewide levels of acetylated histone 3 at lysine 9 (H3K9) at Gfi1 target genes. This led to active expression of genes favoring development of myeloid malignancies [15], which could explain how GFI136N accelerates both AML development and MDS–AML progression. We recently reported that a low level of GFI1 expression, which might mimic the presence of GFI136N on a functional level, accelerates AML progression in different murine models of AML, including one model of MDS [20]. It is not yet clear why altering one amino acid changes the function of GFI1, and initial experiments regarding expression level, stability, ability to induce apoptosis, or interaction with histone deacetylases (HDACs) or lysine-specific demethylase 1 (LSD1) did not reveal any significant differences between GFI136N and GFI136S [15] (unpublished data).
We also examined whether GFI136N could predict response to therapy. To explore this in more detail, we focused on patients that were treated with 5-azacitidine. This treatment is used for patients who are otherwise not fit for allogeneic bone marrow transplantation or as a bridging to a definitive therapy 21, 22, 23. There was no difference between GFI136N heterozygous carriers and GFI136S patients achieving response to treatment with 5-azacytidine (51% of GFI136S homozygous carriers compared with 52% of GFI136N carriers). However, the response to treatment was much shorter in GFI136N carriers than in GFI136S homozygous carriers (Fig. 2E). This observation is not surprising because treatment with hypomethylating agents, such as 5-azacitidine, would not revert the increased levels of H3K9 acetylation and H3K4 methylation seen in cells with a GFI136N allele [15].
The suitability of GFI136N as a prognostic marker should be verified in prospective studies and, if the findings can be confirmed, the status of GFI136N should be determined routinely in MDS patients. Considering the role of GFI1 in myeloid development, GFI136N status could also be of prognostic value for patients with myeloproliferative diseases and chronic myeloid leukemia. Indeed, the frequency of GFI136N is also elevated among patients with chronic myeloid leukemia. In summary, GFI136N could be a useful therapeutic and prognostic marker for myeloid malignancies.
Acknowledgments
CK was supported by a Max Eder Grant from the Deutsche Krebshilfe, Germany, and the IFORES fellowship from the University Clinic Essen, Germany. TM's lab was supported by a grant from the Leukemia and Lymphoma Society (USA). TM holds a Tier 1 Canada Research Chair and grants from the Canadian Institutes of Health Research (CIHR, Grants MOP-84238 and MOP-111011). JB and AP are supported by Bloodwise (UK).
We thank the patients for their consent to participate in the different studies and Saskia Grunwald and Renata Köster for excellent technical assistance.
Footnotes
LB, LCM, HM, JM, and CK contributed equally to this work.
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.exphem.2016.04.001.
Conflict of interest disclosure
Cyrus Khandanpour received travel reimbursement from Amgen and Chugai for attending scientific conferences. Jaroslaw Maciejewski received speaker honoraria from Celgene and Alexion.
Supplementary data
Supplementary Table E1.
Cox survival regression adjusted for IPSSa
| Variable | Wald χ2 | p value | Hazard ratio | 95% CI for hazard ratio |
|
|---|---|---|---|---|---|
| Lower | Upper | ||||
| IPSS | 77.381 | 0.000 | |||
| IPSS low | 13.699 | 0.000 | 0.504 | 0.350 | 0.724 |
| IPSS intermediate 1 | 4.607 | 0.032 | 0.725 | 0.540 | 0.972 |
| IPSS intermediate 2 | 1.586 | 0.208 | 1.229 | 0.892 | 1.694 |
| IPSS high | 32.297 | 0.000 | 3.158 | 2.124 | 4.696 |
| GFI136N | 20.125 | 0.000 | 2.212 | 1.564 | 3.130 |
CI = confidence interval.
IPSS was defined in four different entities as published (for details, see main text).
Supplementary Table E2.
Cox survival regression adjusted for key prognostic factorsa
| Variable | Wald χ2 | P value | Hazard ratio | 95.0% CI for hazard ratio |
|
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Age | 4.761 | 0.029 | 1.010 | 1.001 | 1.019 |
| Blast (% BM) | 7.141 | 0.008 | 1.045 | 1.012 | 1.078 |
| Male | 0.518 | 0.472 | 1.111 | 0.834 | 1.479 |
| IPSS | 5.682 | 0.224 | |||
| IPSS low | 5.081 | 0.024 | 0.571 | 0.351 | 0.930 |
| IPSS intermediate 1 | 2.561 | 0.110 | 0 .728 | 0.494 | 1.074 |
| IPSS intermediate 2 | 0.205 | 0.651 | 0.909 | 0.603 | 1.371 |
| IPSS high | 0.088 | 0.767 | 1.105 | 0.570 | 2.144 |
| Cytogenetic | 12.264 | 0.007 | |||
| Cytogenetic good | 0.115 | 0.735 | 0.894 | 0.466 | 1.713 |
| Cytogenetic intermediate | 1.089 | 0.297 | 0.686 | 0.339 | 1.392 |
| Cytogenetic poor | 1.278 | 0 .258 | 1.469 | 0.754 | 2.862 |
| GFI136N | 10.720 | 0.001 | 2.154 | 1.361 | 3.409 |
CI = confidence interval.
Cox survival regression adjusted for age, blast count, sex, IPSS, cytogenetic finding, and as a last step, presence of GFI136N (see main text for more information).
References
- 1.Phelan J.D., Shroyer N.F., Cook T., Gebelein B., Grimes H.L. Gfi1-cells and circuits: Unraveling transcriptional networks of development and disease. Curr Opin Hematol. 2010;17:300–307. doi: 10.1097/MOH.0b013e32833a06f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vvan der Meer L.T., Jansen J.H., van der Reijden B.A. Gfi1 and Gfi1b: Key regulators of hematopoiesis. Leukemia. 2010;24:1834–1843. doi: 10.1038/leu.2010.195. [DOI] [PubMed] [Google Scholar]
- 3.Hock H., Hamblen M.J., Rooke H.M. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 4.Person R.E., Li F.Q., Duan Z. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet. 2003;34:308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horman S.R., Velu C.S., Chaubey A. Gfi1 integrates progenitor versus granulocytic transcriptional programming. Blood. 2009;113:5466–5475. doi: 10.1182/blood-2008-09-179747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khandanpour C., Thiede C., Valk P.J. A variant allele of growth factor independence 1 (GFI1) is associated with acute myeloid leukemia. Blood. 2010;115:2462–2472. doi: 10.1182/blood-2009-08-239822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hochberg J.C., Miron P.M., Hay B.N. Mosaic tetraploidy and transient GFI1 mutation in a patient with severe chronic neutropenia. Pediatr Blood Cancer. 2008;50:630–632. doi: 10.1002/pbc.21094. [DOI] [PubMed] [Google Scholar]
- 8.Makishima H., Yoshida K., Nguyen N. Somatic SETBP1 mutations in myeloid malignancies. Nat Genet. 2013;45:942–946. doi: 10.1038/ng.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langemeijer S.M., Kuiper R.P., Berends M. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 10.Papaemmanuil E., Gerstung M., Malcovati L. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Germing U., Lauseker M., Hildebrandt B. Survival, prognostic factors and rates of leukemic transformation in 381 untreated patients with MDS and del(5q): A multicenter study. Leukemia. 2012;26:1286–1292. doi: 10.1038/leu.2011.391. [DOI] [PubMed] [Google Scholar]
- 12.Platzbecker U., Al-Ali H.K., Gattermann N. Phase 2 study of oral panobinostat (LBH589) with or without erythropoietin in heavily transfusion-dependent IPSS low or int-1 MDS patients. Leukemia. 2014;28:696–698. doi: 10.1038/leu.2013.325. [DOI] [PubMed] [Google Scholar]
- 13.Schanz J., Tuchler H., Sole F. New comprehensive cytogenetic scoring system for primary myelodysplastic syndromes (MDS) and oligoblastic acute myeloid leukemia after MDS derived from an international database merge. J Clin Oncol. 2012;30:820–829. doi: 10.1200/JCO.2011.35.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris N.L., Jaffe E.S., Diebold J. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: Report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J Clin Oncol. 1999;17:3835–3849. doi: 10.1200/JCO.1999.17.12.3835. [DOI] [PubMed] [Google Scholar]
- 15.Khandanpour C., Krongold J., Schutte J. The human GFI136N variant induces epigenetic changes at the Hoxa9 locus and accelerates K-RAS driven myeloproliferative disorder in mice. Blood. 2012;120:4006–4017. doi: 10.1182/blood-2011-02-334722. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg P.L., Tuechler H., Schanz J. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg P., Cox C., LeBeau M.M. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. [PubMed] [Google Scholar]
- 18.Bejar R., Stevenson K., Abdel-Wahab O. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bejar R., Stevenson K.E., Caughey B.A. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hones J.M., Botezatu L., Helness A. GFI1 as a novel prognostic and therapeutic factor for AML/MDS. Leukemia. 2016 doi: 10.1038/leu.2016.11. [DOI] [PubMed] [Google Scholar]
- 21.Saunthararajah Y. Key clinical observations after 5-azacytidine and decitabine treatment of myelodysplastic syndromes suggest practical solutions for better outcomes. Hematol Am Soc Hematol Educ Program. 2013;2013:511–521. doi: 10.1182/asheducation-2013.1.511. [DOI] [PubMed] [Google Scholar]
- 22.Bejar R., Steensma D.P. Recent developments in myelodysplastic syndromes. Blood. 2014;124:2793–2803. doi: 10.1182/blood-2014-04-522136. [DOI] [PubMed] [Google Scholar]
- 23.Sekeres M.A., Cutler C. How we treat higher-risk myelodysplastic syndromes. Blood. 2014;123:829–836. doi: 10.1182/blood-2013-08-496935. [DOI] [PubMed] [Google Scholar]