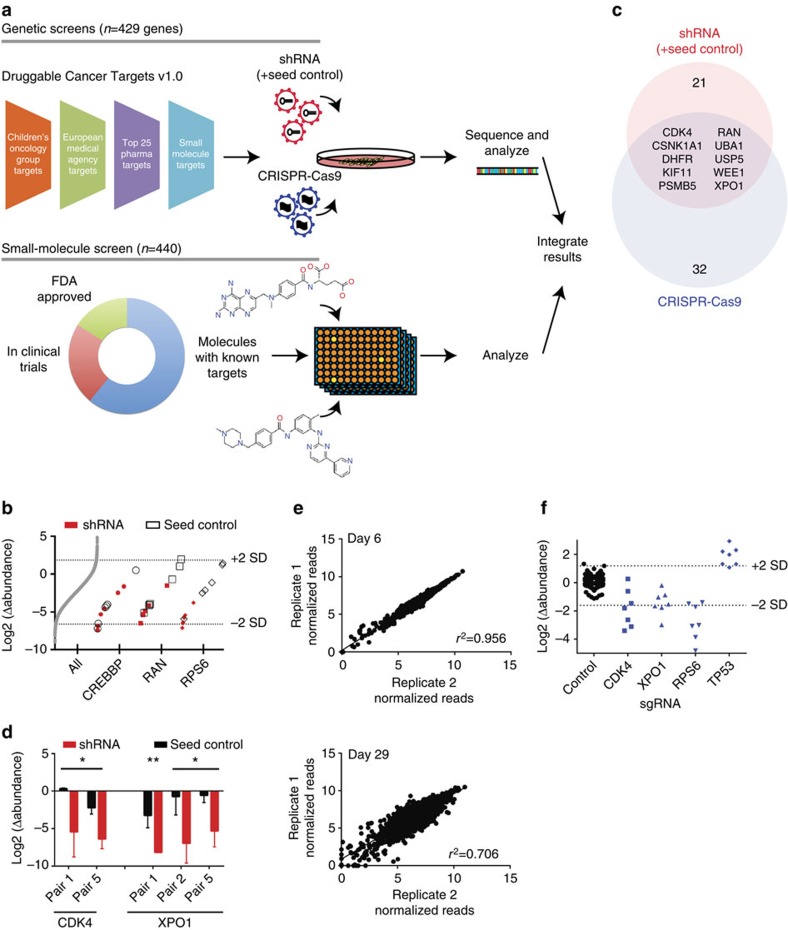
Figure 2. High-throughput functional genomic screens are feasible in an early passaged patient-derived model.
(a) Schema for screens. shRNA and sgRNA libraries were created by compiling targets from the indicated sources and created the Druggable Cancer Targets v1.0 shRNA and sgRNA libraries. In parallel, a compound screen was performed utilizing 440 compounds identified previously9. (b) Using shRNA seed controls to identify off-target shRNAs. Distribution of shRNAs shown in grey. CREBBP was identified as a false positive due to the significant miRNA seed effects (comparing circle outlines, seed controls, to red dots, shRNAs). RAN alternatively was identified as a candidate when accounting for seed effects (square outlines, seed controls, to red squares). A positive control target, RPS6 showed a clear separation between seed controls (diamond outlines) and shRNAs (red diamonds). Each point represents the mean of four biological replicates. (c) Summary of RNAi and CRISPR-Cas9 screens. When comparing candidates from both screens, we found 10 genes that scored in both screens. (d) Detailed comparison of paired shRNAs with shRNA seed controls identified CDK4 and XPO1 in CLF-PED-015-T. Error bars represent mean±s.d. for four independent experiments. (e) Replicates are highly correlated in CLF-PED-015-T following introduction of sgRNAs at days 6 and 29. (f) CRISPR-Cas9 screens in CLF-PED-015-T. Each dot represents the mean of three replicates for a given sgRNA.