Figure 2.
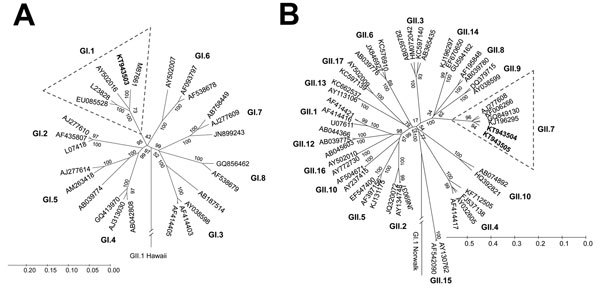
Phylogenetic trees showing GI and GII noroviruses from rhesus macaque fecal samples collected in 2008 (11) and retested in 2015 by using a highly sensitive and specific real-time reverse transcription PCR. GenBank accession numbers or other isolate identifiers are shown. Bold indicates isolates detected in this study. A) GI noroviruses share 100% nucleotide homology with the prototype Norwalk virus GI.1 strain (M87661). B) GII noroviruses group with GII.7 human noroviruses. Three of the 4 GII norovirus open reading frame (ORF) 2 sequences obtained in this study were identical. Only nonidentical sequences are shown. Phylogenetic trees were constructed on the basis of alignments of full length ORF2 nucleotide sequences, by using the unweighted pair group method with arithmetic mean and the neighbor-joining clustering methods of the Molecular Evolutionary Genetics Analysis (MEGA version 6.1; http://mega.software.informer.com/6.1/) software with Jukes-Cantor distance calculations. The confidence values of the internal nodes were obtained by performing 1,000 bootstrap analyses. Scale bars represent nucleotide substitutions per site.
