Figure.
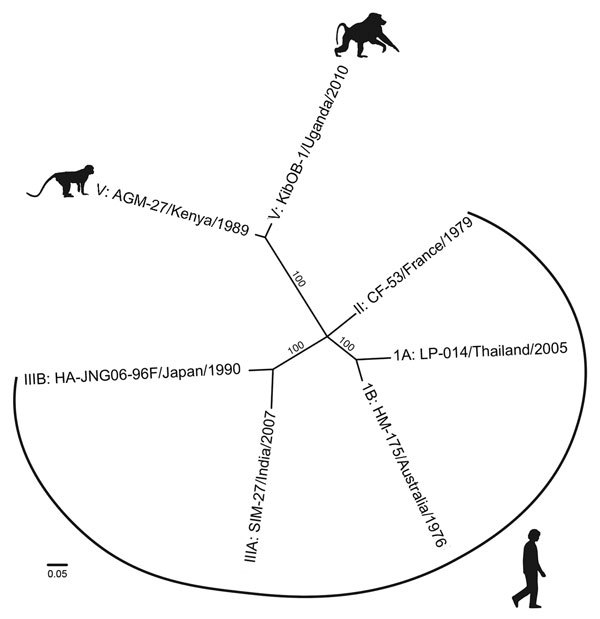
Whole-genome phylogenetic reconstruction of representative HAVs. HAVs are grouped into 6 genotypes based on 168 bp of the C-terminal extension of the viral protein 1 gene. Baboon HAV detected in Kibale National Park, Uganda, in 2010 and 2014 (GenBank accession number KT819575) clusters with AGM-27 (3), previously the sole member of genotype V. jModeltest 2 (http://jmodeltest.org) was used to find the best-fit evolutionary model for the data, after which the maximum-likelihood tree was estimated using the heuristic search method in PAUP* (http://paup.csit.fsu.edu), with starting trees obtained by neighbor-joining, random stepwise addition, and branch swapping by tree-bisection reconnection and starting branch lengths obtained using Rogers-Swofford approximation. Bootstrap values were derived from 1,000 replicates of the heuristic search; only values >50% are shown. GenBank accession nos.: IA, EF207320; IB, M14707; II, AY644676; IIIA, FJ227135; IIIB, AB258387; V, D00924). HAV, hepatitis A virus. Scale bar indicates substitutions per site.
