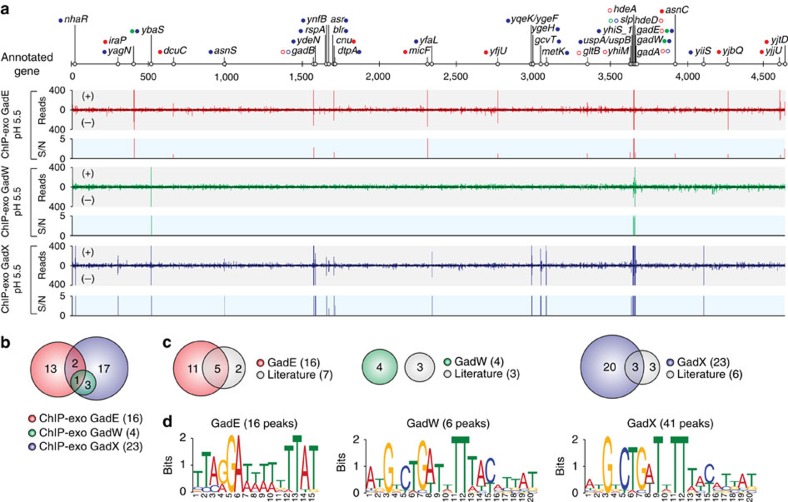
Figure 1. Genome-wide landscape of GadEWX-binding sites.
(a) An overview of GadEWX-binding profiles across the E. coli genome (KiloBase) at mid-exponential growth phase (OD600=0.3) under acidic stress (pH 5.5). Open and closed dots indicate previously known and newly found GadEWX-binding sites, respectively. S/N denotes signal-to-noise ratio. (+) and (−) indicate reads mapped on forward and reverse strands, respectively. Binding peaks that overlap with Mock-IP signal were eliminated. Red, GadE; Green, GadW; Blue, GadX. (b) Overlaps between GadEWX-binding sites under the low-pH condition. (c) Comparison of the GadEWX binding sites obtained from this study (ChIP-exo) with the known binding sites from the literature. (d) Sequence logo representations of the GadEWX-DNA-binding motifs.