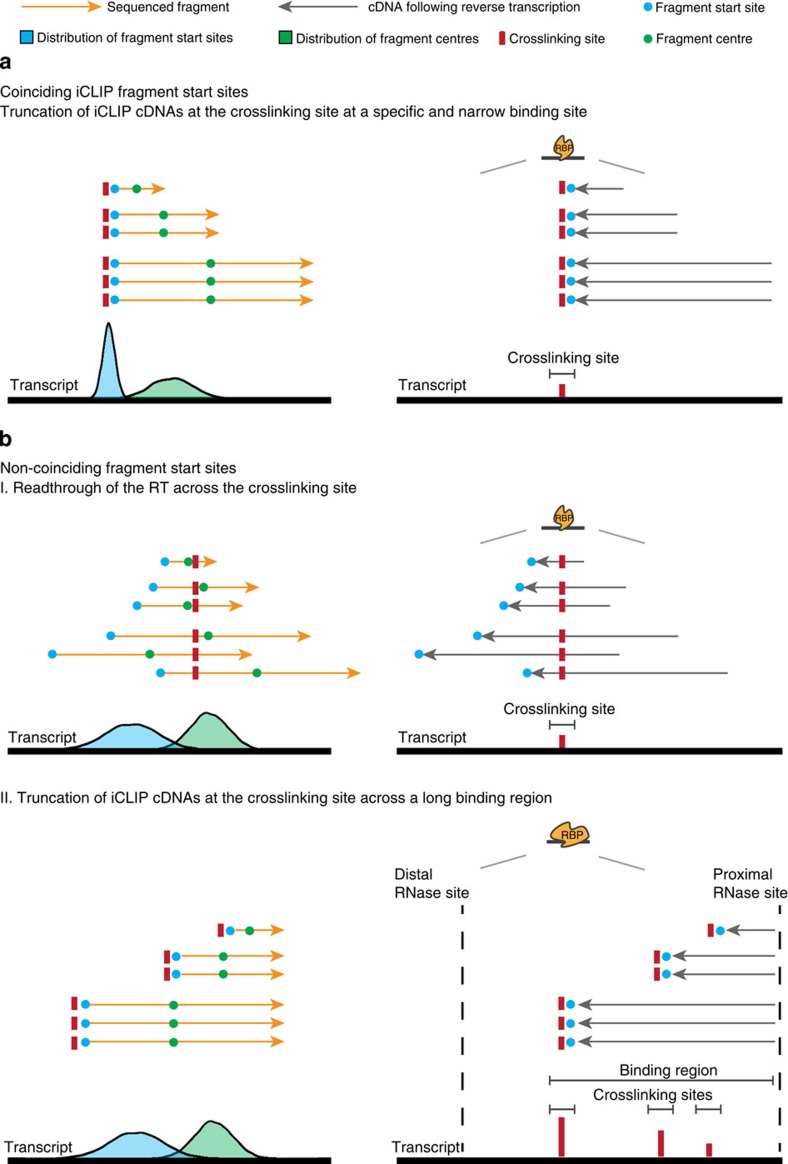
Figure 7. Schematic representations of potential mechanisms at the reverse transcription step in iCLIP.
(a) Coinciding iCLIP fragment start sites. The crosslinking site can be defined in a manner independent of fragment length as the nucleotide preceding the iCLIP fragment, if RT synthesis is dominated by truncation. The distribution of other reference points within the iCLIP fragment such as the centre will be much broader when compared with the distribution of the start sites, because of the different lengths of the sequenced fragments. (b) Non-coinciding iCLIP fragment start sites. (I) A read-through mechanism or (II) long crosslinking regions are plausible explanations for a broad distribution of iCLIP fragment start sites. In case of read-through, the centre positions of the iCLIP fragments will be closer to the distribution of the crosslinking sites than the start sites of the iCLIP fragments. In case the protein binds and crosslinks across a long stretch of RNA, the iCLIP fragments of different lengths may sample different crosslinking sites. The overlap of iCLIP fragment ends suggests that the protein protects the RNA from RNase digestion. In case of predominantly non-overlapping start sites the use of the fragment start site as a reference point for mapping binding sites is dependent on the fragment length distribution (see Supplementary Fig. 1). The use of the centre position will reduce this length-dependency and improves the assignment of binding sites due to the increased signal of overlapping centres compared with start sites of the fragments.