Figure 4.
Sas-4-Thr200 Is Required for Centriole Conversion
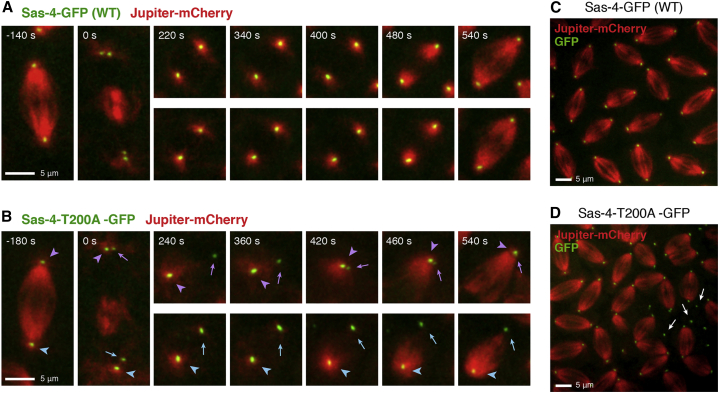
(A and B) Time-lapse images show the localization of Jupiter-mCherry (MT marker, red) in living embryos expressing WT Sas-4-GFP (A) or Sas-4-T200A-GFP (B) from injected mRNA (green, as indicated). (A) In embryos expressing WT Sas-4-GFP, centrioles separate at the end of mitosis (t = 0); both centrioles form centrosomes that nucleate robust MT arrays throughout S phase (t = 220–400 s) and later organize the mitotic spindle poles (t = 540 s). (B) In embryos expressing Sas-4-T200A-GFP, the old centrioles (arrowheads) organize a centrosome that nucleates MTs, but new centrioles (arrows) are unable to efficiently convert into functional MT-organizing centers (t = 240–540 s), leading to spindle abnormalities during mitosis (t = 540 s).
(C and D) Images show metaphase-stage living embryos expressing WT Sas-4-GFP (C) or Sas-4-T200A-GFP (D) from injected mRNA (green, as indicated). Jupiter-mCherry (red) localization shows the mitotic spindles. Note that embryos expressing Sas-4-T200A-GFP contain multiple “unconverted” centrioles that do not participate in spindle formation; in these embryos abnormal, conjoined spindles are formed by the old centrosomes while the new centrioles float nearby in the cytoplasm (white arrows in D). Such spindle defects were observed in six of nine embryos injected with Sas-4-T200A-GFP, but in none of three embryos injected with WT Sas-4-GFP.
Scale bars, 5 μm.