Extended Data Figure 5. Motala haplotypes carrying the derived, selected EDAR allele.
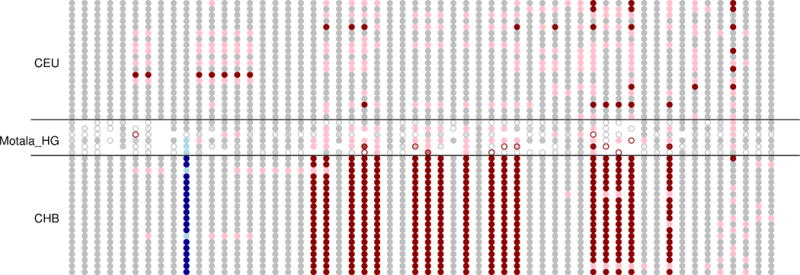
This figure compares the genotypes at all sites within 150kb of rs3827760 (in blue) for the 6 Motala samples and 20 randomly chosen CHB (Chinese from Beijing) and CEU (Utah residents with northern and western European ancestry) samples. Each row is a sample and each column is a SNP. Grey means homozygous for the major (in CEU) allele. Pink denotes heterozygous and red homozygous for the other allele. For the Motala samples, an open circle means that there is only a single sequence otherwise the circle is colored according to the number of sequences observed. Three of the Motala samples are heterozygous for rs3827760 and the derived allele lies on the same haplotype background as in present-day East Asians. The only other ancient samples with evidence of the derived EDAR allele in this dataset are two Afanasievo samples dating to 3300-3000 BCE, and one Scythian dating to 400-200 BCE (not shown).
