FIGURE 2.
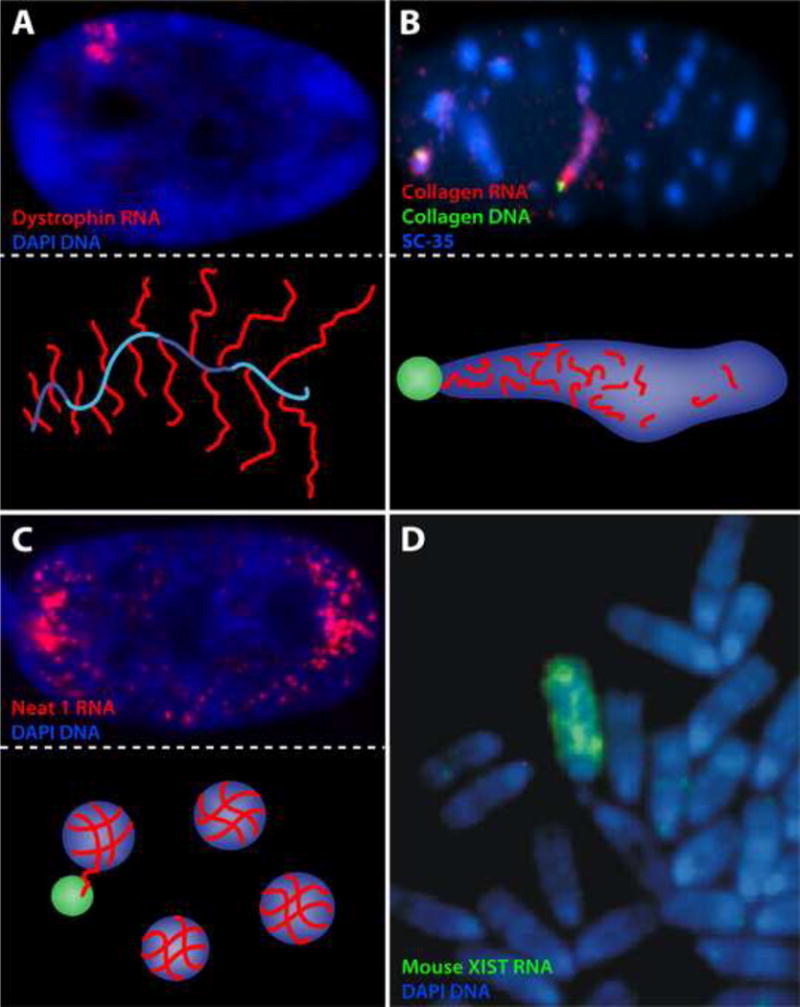
A) Photo: The large (2MB) dystrophin gene produces a substantial accumulation of nascent transcripts localized over chromatin. Illustration: Other analyses [19] showed this was a “tree” of multiple transcripts (red) in synthesis on the large gene (blue), producing what looks like a small fir tree (which can be often seen by electron microscope). B) Photo: Collagen pre/mRNAs (red), which require processing of 50 introns in a relatively small transcription unit, produce a large post-transcriptional accumulation in non-chromatin Nuclear Speckles (visualized using the spliceosome assembly factor SC35), which is adjacent to the Collagen genes (green) [18]. Illustration: Collagen pre-mRNA (red) emanates from the gene (green) and is processed in the Nuclear Speckle (blue). C) Photo: NEAT1 is an architectural-RNA that is required to form the underpinnings of non-chromatin nuclear structures termed paraspeckles, enriched for specific proteins. Illustration: The paraspeckles (blue) form on NEAT1 RNA (red) transcribed at the gene locus (green) but then move into the nucleoplasm [21]. D) Mouse Xist RNA in some mouse cell types is retained long enough to be glimpsed on mitotic chromosomes before it detaches, and can be seen to paint the whole Xi except the centromere.
