FIGURE 4.
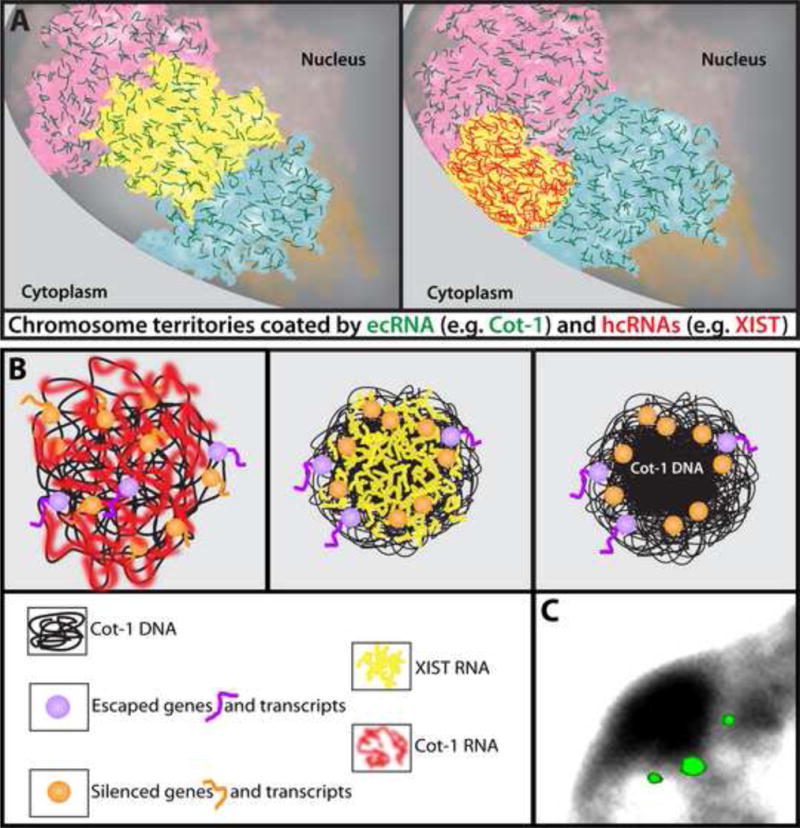
A) Model for the widespread presence of RNA bound throughout much of interphase chromatin, with two broad types of “chromosomal RNAs” (cRNAs): euchromatin-associated chromosomal RNAs (ecRNAs, green) present on active chromatin, such as Cot-1 ecRNAs, and heterochromatin-associated chromosomal RNAs (hcRNAs), such as XIST RNA (red). Left: All three chromosomes (pink, yellow and blue) are active and coated with ecRNA. Right: When XIST hcRNA coats the previously active yellow chromosome, ecRNAs are lost, and this potentially contributes to chromosome condensation. Whether there are other hcRNAs associated with other facultative heterochromatin at the nuclear periphery remains to be determined (note: Cot-1 ecRNAs are not found in silenced chromosomal regions up against the nuclear periphery). B) Left: Cot-1 ecRNA (red) is expressed from active chromosomes (e.g. Xa) which may help maintain an open chromatin state. Middle: When XIST RNA (yellow) first coats the X-chromosome, Cot-1 ecRNA is silenced, which may facilitate the compaction of Cot-1 DNA into the core of the chromosome territory forming the BB. Right: Genes position at the periphery of this condensed repeat-rich core to facilitate efficient silencing, while genes that escape silencing may be more peripheral. C) Simultaneous hybridization to three silenced genes on the inactive X-chromosome position just outside the condensed BB similar to the illustration at right above [12].
