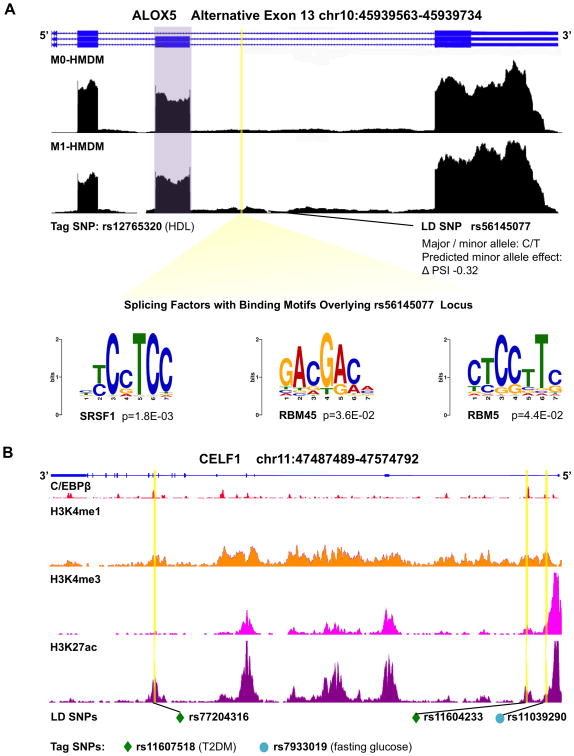
Figure 4. Cardiometabolic trait-associated variants within genomic loci of alternatively spliced genes and splicing factors.
A, Genome browser view of ALOX5 RNA-seq wiggle tracks showing more exon 13 (purple) inclusion in M1-HMDMs, with a SNP, in LD with a tag SNP for HDL-C, predicted to disrupt the splicing code to decrease exon inclusion by 32%. The LD SNP falls within the binding sites for splicing factors SRSF1, RBM45, and RBM5. B, Genome browser view of splicing factor gene CELF1: RefSeq annotation in blue, C/EBPβ ChIP-seq peaks,58 and histone tail modification marks on ChIP-seq.81 Yellow highlights indicate positions of SNPs in LD with tag SNPs for T2DM (green diamond) and fasting glucose (teal circle), notably overlapping C/EBPβ binding sites. HMDMs = human monocyte derived macrophages; SNP = single nucleotide polymorphism; LD = linkage disequilibrium; ChIP-seq = chromatin immunoprecipitation sequencing; T2DM = type 2 diabetes mellitus.