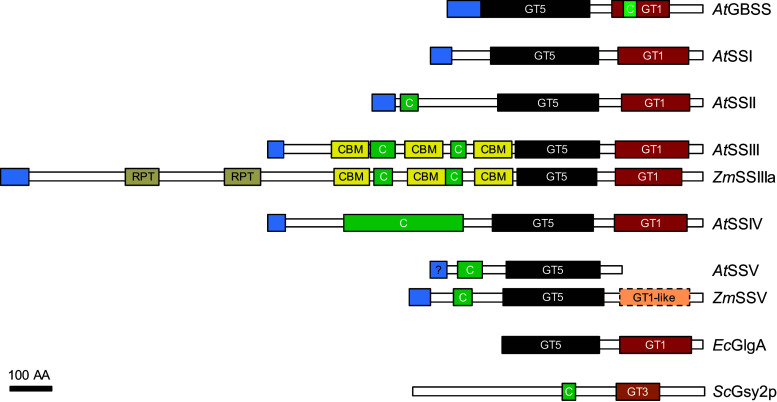
Fig. 2.
The domain structure of starch synthase (SS) classes. SSs from Arabidopsis (At) compared with glycogen synthases from E. coli (Ec) and budding yeast (Sc). Maize (Zm) SSIIIa and SSV are included as they differ in their structures compared with the Arabidopsis orthologs. Shown are plastidial transit peptides (N-terminal blue boxes), internal repeats (gray boxes, RPT), carbohydrate-binding modules of family 25 (yellow boxes, CBM), coiled-coil domains (green boxes, C), glycosyltransferase-5 domains (black boxes, GT5), glycosyltransferase-1 domains (red boxes, GT1) and a glycosyltransferase-3 domain (orange box, GT3). Transit peptides were predicted with ChloroP [285], coiled-coil motifs with Paircoil2 ([286]; p value < 0.05, 21 amino acids minimal length) and all other motifs with SMART. Note that the domain length and annotation depend on the database queried. For example, the GT3 domain of ScGsy2P is identified as a GT1 domain by SMART and was manually re-assigned as GT3 here [287]. Whereas ScGsy2p is a GT3 family glycosyltransferase and uses UDPglucose as substrate, all other shown synthases are GT5 family glycosyltransferases and use ADPglucose as substrate. The N-terminal regions of SSIII containing the coiled-coil motifs and CBMs are highly conserved among various orthologs, but further contain internal repeats in some cases (e.g., in barley and wheat SSIIIa). Orthologs of SSII often showed weaker or no coiled-coil predictions. AtSSV has a weakly predicted chloroplast transit peptide, but lacks the GT1 domain and is of unknown function. SSV from most other species, however, have a C-terminal extension including a stretch that was designated as a putative GT1-like domain [60]. Bar 100 amino acids (AA)