Abstract
Accumulation of sterols in endoplasmic reticulum membranes stimulates the ubiquitination of 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR), which catalyzes a rate-limiting step in synthesis of cholesterol. This ubiquitination marks HMGCR for proteasome-mediated degradation and constitutes one of several mechanisms for feedback control of cholesterol synthesis. Mechanisms for sterol-accelerated ubiquitination and degradation of HMGCR have been elucidated through the study of cultured mammalian cells. However, the extent to which these reactions modulate HMGCR and contribute to control of cholesterol metabolism in whole animals is unknown. Here, we examine transgenic mice expressing in the liver the membrane domain of HMGCR (HMGCR (TM1–8)), a region necessary and sufficient for sterol-accelerated degradation, and knock-in mice in which endogenous HMGCR harbors mutations that prevent sterol-induced ubiquitination. Characterization of transgenic mice revealed that HMGCR (TM1–8) is appropriately regulated in the liver of mice fed a high cholesterol diet or chow diet supplemented with the HMGCR inhibitor lovastatin. Ubiquitination-resistant HMGCR protein accumulates in the liver and other tissues disproportionately to its mRNA, indicating that sterol-accelerated degradation significantly contributes to feedback regulation of HMGCR in vivo. Results of these studies demonstrate that HMGCR is subjected to sterol-accelerated degradation in the liver through mechanisms similar to those established in cultured cells. Moreover, these studies designate sterol-accelerated degradation of HMGCR as a potential therapeutic target for prevention of atherosclerosis and associated cardiovascular disease.
Keywords: cholesterol metabolism, endoplasmic reticulum (ER), endoplasmic-reticulum-associated protein degradation (ERAD), isoprenoid, lipid metabolism
Introduction
3-Hydroxy-3-methylglutaryl coenzyme A (HMG-CoA)2 reductase (HMGCR) is a polytopic protein of endoplasmic reticulum (ER) membranes that catalyzes conversion of HMG-CoA to mevalonate, a rate-limiting reaction in synthesis of cholesterol as well as nonsterol isoprenoids, such as dolichol, ubiquinone, farnesyl pyrophosphate, and geranylgeranyl pyrophosphate (1). Multiple feedback mechanisms converge on HMGCR to ensure that essential nonsterol isoprenoids are continuously synthesized while avoiding overaccumulation of cholesterol and other sterols (2). Recent work indicates additional control points in the cholesterol biosynthetic pathway beyond HMGCR (3). One mechanism for feedback control of HMGCR involves its accelerated degradation from ER membranes (4, 5). This degradation is initiated by intracellular accumulation of sterols, which causes HMGCR to bind to ER membrane proteins called Insig-1 and Insig-2 (6). Insig binding is mediated entirely by the membrane domain of HMGCR, which contains eight transmembrane helices and precedes a cytosolic catalytic domain (7, 8). Insig-associated ubiquitin ligases facilitate ubiquitination of cytosolically exposed lysine residues in the membrane domain of HMGCR (9–11), marking the enzyme for extraction across ER membranes and dislocation into the cytosol for proteasomal degradation (12, 13).
A second mechanism for feedback regulation of HMGCR involves sterol-induced binding of Insigs to Scap, another polytopic protein of ER membranes (14). Scap associates with membrane-bound sterol regulatory element-binding proteins (SREBPs) that modulate transcription of genes encoding HMGCR and other cholesterol biosynthetic enzymes (15). In sterol-depleted cells, Scap facilitates transport of SREBPs from ER to Golgi, where transcriptionally active SREBP fragments are proteolytically released from membranes. These fragments migrate to the nucleus and activate target gene transcription. Excess sterols cause Insigs to bind to Scap, which inhibits transport of Scap-SREBP complexes from ER to Golgi. In the absence of transport, proteolytic activation of SREBPs does not occur, and as a result, expression of mRNAs encoding SREBP target genes and cholesterol synthesis declines.
Transcriptional control of HMGCR governed by Scap-SREBP has been extensively studied in livers of transgenic and knock-out mice (16). In contrast, little is known about degradative control of hepatic HMGCR, due in part to the inability to directly measure the parameter in vivo (17–19). In Insig-deficient mouse livers, HMGCR protein accumulates disproportionately to its mRNA (20), which is probably due to the combination of defects in transcriptional and degradative control of HMGCR. However, the extent to which these mechanisms individually contribute to HMGCR regulation in Insig-deficient livers remains to be determined.
To clarify the role of accelerated degradation in feedback regulation of HMGCR, we generated two lines of mice: 1) transgenic mice expressing in the liver the membrane domain of HMGCR, which is necessary and sufficient for Insig-mediated, sterol-accelerated degradation (6), and 2) knock-in mice harboring mutations in the endogenous HMGCR gene that change lysine residues 89 and 248 to arginine. These mutations abolish sterol-induced ubiquitination and subsequent degradation of HMGCR in cultured cells (21). Characterization of these mice reveals that sterols directly modulate degradation of HMGCR in the liver through mechanisms similar to those previously established in cultured cells. Moreover, our results indicate that sterol-accelerated degradation contributes to regulation of HMGCR and cholesterol metabolism in vivo.
Experimental Procedures
Determination of Metabolic Parameters
Blood was drawn from the vena cava after WT, knock-in, and transgenic mice were anesthetized in a bell jar atmosphere containing isoflurane. Plasma was immediately separated and stored at −80 °C until use. Commercial kits from Wako Chemicals USA, Inc. (Richmond, VA) were used to measure non-esterified fatty acids (HR Series NEFA-HR (2)). Levels of plasma and liver cholesterol and triglycerides were measured using the Infinity total cholesterol and Infinity triglyceride kits (ThermoFisher Scientific). The level of plasma insulin in WT, knock-in, and transgenic mice was measured using an ELISA kit from Crystal Chem Inc. (Downers Grove, IL) (catalogue no. 90080). Plasma glucose was measured with a Contour Glucometer (Bayer) from the tail nick of conscious mice.
Generation of hmgcr Knock-in and Tg-HMGCR (TM1–8) Mice
HMGCR knock-in mice, which harbor mutations of lysines 89 (AAG) and 248 (AAA) to arginine (AGG and AGA, respectively), were generated by the Gene Targeting and Transgenic Facility at the Howard Hughes Medical Institute Janelia Research Campus (Ashburn, VA). Genotyping was carried out as described below.
To generate Tg-HMGCR (TM1–8) mice, we used the pLiv-11 vector containing the constitutive human apoE gene promoter and its hepatic control region (22). Transgenic plasmid pLiv-11-HMGCR (TM1–8) was generated by cloning a cDNA fragment encoding the membrane domain (amino acids 1–346) of hamster HMGCR followed by three T7 epitopes (6) into MluI-ClaI sites of pLiv-11. The SalI-SpeI fragment of pLiv-11-HMGCR (TM1–8) was isolated and injected into fertilized eggs to generate transgenic mice by the Transgenic Core Facility at the University of Texas Southwestern Medical Center. Founder mice were identified, positive founders were bred to C57BL/6J mice, and three lines of Tg-HMGCR (TM1–8) mice were established. One line expressing a moderate level of HMGCR (TM1–8) in the liver was chosen for further study.
WT and hmgcr knock-in mice (hmgcrWT/Ki and hmgcrKi/Ki) littermates were obtained for experiments from intercrosses of hmgcrWT/Ki heterozygous male and female mice, all of which were hybrids of C57BL/6J and 129Sv/Ev strains. Tg-HMGCR (TM1–8) mice were maintained as hemizygotes by breeding with WT C57BL/6J mice. Mice were housed in colony cages with a 12-h light/12-h dark cycle and fed Teklad Mouse/Rat Diet 2016 from Harlan Teklad (Madison, WI). For experiments, non-transgenic WT littermates were used as controls for the transgenic mice. All animal experiments were performed with the approval of the institutional animal care and use committee at the University of Texas Southwestern Medical Center at Dallas.
Genotyping of Tg-HMGCR (TM1–8) and hmgcr Knock-in Mice
Genomic DNA was extracted from tails of Tg-HMGCR (TM1–8) mice using a DNeasy blood and tissue kit (Qiagen, Venlo, Netherlands) according to the manufacturer's protocol. To genotype the transgenic animals, genomic DNA was amplified with the following primers: forward, 5′-GCCCTAAGTTCAAACTCTCAGGATGAAG-3′; reverse, 5′-GGGCCCTCTAGATCACATATTAATTAAACCC-3′. A primer set targeting the mouse Hbb-b1 gene (forward, 5′-CCAATCTGCTCACACAGGATAGAGAGGGCAGG-3′; reverse, 5′-CCTTGAGGCTGTCCAAGTGATTCAGGCCATCG-3′) was used as a positive control. To genotype the hmgcr knock-in animals, genomic DNA from tails was used for PCR with the following primers: set A forward, 5′-GTCCATGAACATGTTCACCG-3′; set A reverse, 5′-CAGCACGTCCTATTGGCAGA-3′; set B forward, 5′-TCGGTGATGTTCCAGTCTTC-3′; set B reverse, 5′-GGTGGCAAACACCTTGTATC-3′. Genotypes of the knock-in animals were further verified by the DNA Sanger Sequencing Core Facility at the University of Texas Southwestern Medical Center using the sequencing primers (K89R, 5′-GTATCACTGAGGCCTCTCAT-3′; K248R, 5′-AGTGCCCACTTCCTTCGTAG-3′) after amplification of the genomic DNA with the following primers: K89R forward, 5′-TTCTCTGCCAATAGGACGTG-3′; K89R reverse, 5′-TAGAAGAGCACTGCCACGTT-3′; K248R forward, 5′-AGTAGTACTTCCCATGCTGC-3′; K248R reverse, 5′-GAAGACTGGAACATCACCGA-3′.
Diet Studies
For the cholesterol feeding studies, mice were fed a chow diet (Teklad Mouse/Rat 2016, 0% cholesterol) or chow diet supplemented with 0.05, 0.2, or 2% cholesterol for 5 days before study. For lovastatin feeding studies, mice were fed Teklad Mouse/Rat diet 7002 (Harlan Teklad Premier Laboratory Diets, Madison, WI) or the identical diet supplemented with 0.02, 0.06, or 0.2% lovastatin (Abblis Chemicals LLC, Houston, TX). For fasting and refeeding experiments, mice were divided into three groups: nonfasted, fasted, and refed. The nonfasted group was fed ad libitum, the fasted group was fasted for 12 h, and the refed group was fasted for 12 h and then refed a high carbohydrate/low fat diet (MP Biomedicals, Santa Ana, CA) for 12 h before study. The starting times for the experiments were staggered such that all mice were sacrificed at the same time, which was at the end of the dark cycle. Liver cholesterol content of WT and Tg-HMGCR (TM1–8) mice fed cholesterol in Fig. 2 were as follows: WT, chow (2.3 ± 0.2 mg/g), 0.05% cholesterol (2.3 ± 0.2 mg/g), 0.2% cholesterol (4.5 ± 0.2 mg/g), and 2% cholesterol (3.6 ± 0.4 mg/g); Tg-HMGCR (TM1–8), chow (2.1 ± 0.3 mg/g), 0.05% cholesterol (2.5 ± 0.3 mg/g), 0.2% cholesterol (3.6 ± 0.3 mg/g), and 2% cholesterol (4.3 ± 0.2 mg/g). Liver cholesterol content of WT and Tg-HMGCR (TM1–8) mice fed lovastatin in Fig. 3 were as follows: WT, chow (2.0 ± 0.1 mg/g), 0.2% lovastatin (2.0 ± 0.2 mg/g); Tg-HMGCR (TM1–8), chow (1.4 ± 0.3 mg/g), 0.2% lovastatin (2.1 ± 0.2 mg/g). Values represent mean ± S.E. of data from four mice.
FIGURE 2.
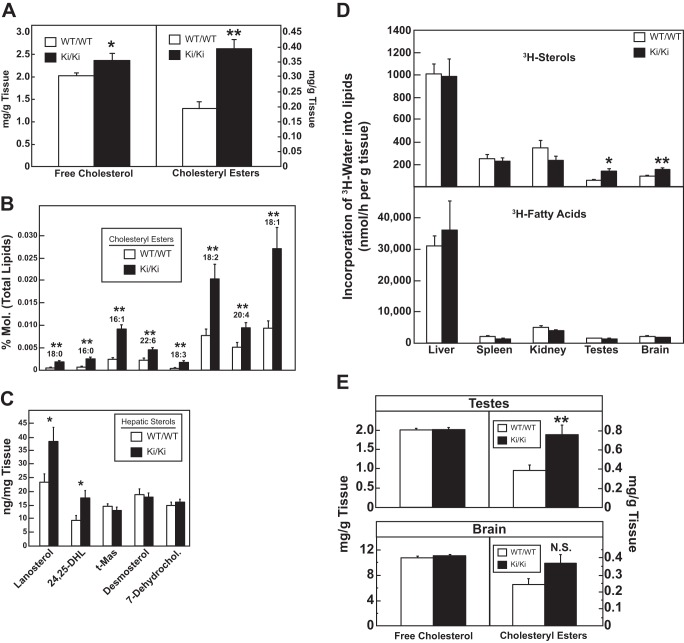
Analysis of cholesteryl esters, cholesterol biosynthetic intermediates, and sterol synthesis in WT and hmgcr knock-in mice. Male mice (6–8 weeks of age, 5–6 mice/group) were fed ad libitum a chow diet before study. Livers (A–C) or brains and testes (E) were collected, and the amounts of free cholesterol, cholesteryl esters, and cholesterol biosynthetic intermediates were determined by a colorimetric assay (A and E) or by LC-MS/MS (C) as described under “Experimental Procedures.” Error bars, S.E. D, male mice (6–8 weeks of age, 5–6 mice/group) were fed a chow diet ad libitum and injected intraperitoneally with 3H-labeled water (50 mCi in 0.2 ml of isotonic saline). One hour later, tissues were removed for measurement of 3H-labeled fatty acids and digitonin-precipitable sterols. Each bar represents the mean ± S.E. of the values from 5 or 6 mice. 24,25-DHL, 24,25-dihydrolanosterol; t-MAS, testis-specific meiosis-activating sterol; 7-Dehydrochol., 7-dehydrocholesterol. *, p < 0.05; **, p < 0.01; N.S., not significant.
FIGURE 3.
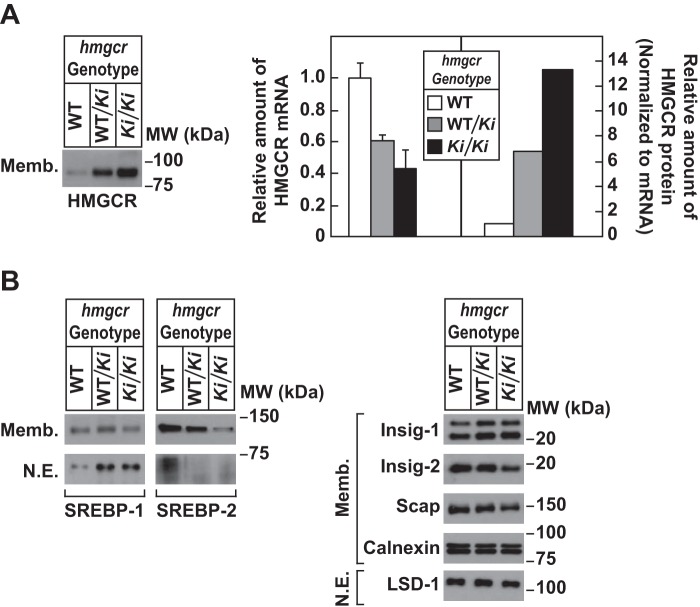
Levels of endogenous HMGCR in livers of WT and hmgcr knock-in mice. Thirteen-week-old male WT, hmgcrWT/Ki, and hmgcrKi/Ki littermates (4 mice/group) were fed an ad libitum chow diet before study. Livers of mice were subjected to subcellular fractionation as described under “Experimental Procedures.” Aliquots of resulting membrane (Memb., 30 μg of protein/lane) and nuclear extract (N.E., 20–50 μg of protein/lane) fractions for each group were pooled and subjected to immunoblot analysis using antibodies against endogenous HMGCR, SREBP-1, SREBP-2, Insig-1, Insig-2, Scap, calnexin, and LSD-1. Although shown in separate panels, Scap and calnexin serve as loading controls for the HMGCR immunoblot. For mRNA analysis (A), equal amounts of RNA from individual mice were subjected to quantitative real-time RT-PCR using primers against the HMGCR mRNA and apoB mRNA as an invariant control. The relative amount of HMGCR protein in hmgcr knock-in mice was determined by quantifying the band corresponding to HMGCR using Image J software and normalizing it to the amount of HMGCR mRNA. Error bars, S.E.
Quantitative Real-time PCR
Total RNA was prepared from mouse tissues using the RNA STAT-60 kit (TEL-TEST “B”, Friendswood, TX). Equal amounts of RNA from individual mice were treated with DNase I (DNA-freeTM, Ambion/Life Technologies, Inc.). First strand cDNA was synthesized from 2 μg of DNase I-treated total RNA with random hexamer primers using TaqMan reverse transcription reagents (Applied Biosystems/Roche Applied Science). Specific primers for each gene were designed using Primer Express software (Life Technologies). The real-time RT-PCR was set up in a final volume of 20 μl containing 20 ng of reverse-transcribed total RNA, 167 nm forward and reverse primers, and 10 μl of 2× SYBR Green PCR Master Mix (Life Technologies). PCRs were done in triplicate. The relative amount of all mRNAs was calculated using the comparative threshold cycle (CT) method. Mouse apoB mRNA was used as the invariant controls. The primers for real-time RT-PCR were described previously (20).
Subcellular Fractionation and Immunoblot Analysis
Approximately 50 mg of frozen liver was homogenized in 350 μl of buffer (10 mm HEPES-KOH, pH 7.6, 1.5 mm MgCl2, 10 mm KCl, 5 mm EDTA, 5 mm EGTA, and 250 mm sucrose) supplemented with a protease inhibitor mixture consisting of 0.1 mm leupeptin, 5 mm dithiothreitol, 1 mm PMSF, 0.5 mm Pefabloc, 5 μg/ml pepstatin A, 25 μg/ml N-acetyl-Leu-Leu-norleucinal, and 10 μg/ml aprotinin. The homogenates were then passed through a 22-gauge needle 10–15 times and subjected to centrifugation at 1000 × g for 5 min at 4 °C. The 1,000 × g pellet was resuspended in 500 μl of buffer (20 mm HEPES-KOH, pH 7.6, 2.5% (v/v) glycerol, 0.42 m NaCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA) supplemented with the protease inhibitor mixture, rotated for 30 min at 4 °C, and centrifuged at 100,000 × g for 30 min at 4 °C. The supernatant from this spin was precipitated with 1.5 ml of cold acetone at −20 °C for at least 30 min; the precipitated material was collected by centrifugation, resuspended in SDS lysis buffer (10 mm Tris-HCl, pH 6.8, 1% (w/v) SDS, 100 mm NaCl, 1 mm EDTA, and 1 mm EGTA), and designated the nuclear extract fraction. The postnuclear supernatant from the original spin was used to prepare the membrane fraction by centrifugation at 100,000 × g for 30 min at 4 °C. Each membrane fraction was resuspended in 100 μl of SDS lysis buffer.
Protein concentrations of nuclear extract and membrane fractions were measured using the BCA kit (ThermoFisher Scientific). Before SDS-PAGE, aliquots of the nuclear extract fractions were mixed with 4× SDS-PAGE loading buffer to achieve a final concentration of 1×. Aliquots of the membrane fractions were mixed with an equal volume of buffer containing 62.5 mm Tris-HCl, pH 6.8, 15% (w/v) SDS, 8 m urea, 10% (v/v) glycerol, and 100 mm DTT, after which 4× SDS loading buffer was added to a final concentration of 1×. Nuclear extract fractions were boiled for 5 min, and membrane fractions were incubated for 20 min at 37 °C before SDS-PAGE. After SDS-PAGE, proteins were transferred to Hybond C-Extra nitrocellulose filters (GE Healthcare). The filters were incubated with the antibodies described below and in the figure legends. Bound antibodies were visualized with peroxidase-conjugated, affinity-purified donkey anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) using the SuperSignal CL-HRP substrate system (ThermoFisher Scientific) according to the manufacturer's instructions. Gels were calibrated with prestained molecular mass markers (Bio-Rad). Filters were exposed to film at room temperature. Antibodies used for immunoblotting to detect mouse SREBP-1 (rabbit polyclonal IgG-211C), SREBP-2 (rabbit monoclonal IgG-22D5), Insig-1 (rabbit polyclonal anti-Insig-1 antiserum), Insig-2 (rabbit polyclonal IgG-940), HMGCR (IgG-839c), gp78 (rabbit polyclonal IgG-740F), and Scap (IgG-R139) were described previously (10, 20, 23). Mouse monoclonal anti-T7 tag IgG was obtained from EMD Biosciences (San Diego, CA). Rabbit polyclonal anti-calnexin IgG was purchased from Novus Biologicals (Littleton, CO). Rabbit polyclonal anti-LSD1 IgG was obtained from Cell Signaling (Beverly, MA). All antibodies were used at a final concentration of 1–5 μg/ml; the anti-Insig-1 antiserum was used at a dilution of 1:1000.
Ubiquitination of Hepatic HMGCR
Approximately 35 mg of frozen liver was homogenized in 1 ml of PBS containing 1% Nonidet P-40, 1% deoxycholic acid, 5 mm EDTA, 5 mm EGTA, 0.1 mm leupeptin, 10 mm N-ethylmaleimide, and the protease inhibitor mixture and subjected to centrifugation at 16,000 × g for 15 min at 4 °C. Immunoprecipitation of the clarified lysates was carried out with polyclonal antibodies against the catalytic domain of human HMGCR as described previously (21). Aliquots of the immunoprecipitates were subjected to SDS-PAGE followed by immunoblot analysis with mouse monoclonal antibodies IgG-A9 (against HMGCR), IgG-P4D1 (against ubiquitin), and IgG-819 against UBXD8 (24).
Lipid Analysis
Sterol biosynthetic intermediates were measured using LC-MS/MS according to the method of McDonald et al. (25, 26). Briefly, sterols were isolated on an LC gradient (Shimadzu LC-20) and detected using the multiple-reaction monitoring pair on a triple quadrupole MS (ABSciex 4000 q-TRAP) and quantified against authentic sterol standards (Avanti Polar Lipids, Alabaster, AL).
Cholesteryl esters were measured by directly infusing a Bligh-Dyer extract into a triple-TOF MS (ABSciex 5600+), commonly known as “shotgun lipidomics.” Cholesteryl esters were identified based on their characteristic 369-Da fragment ion, and the fatty acid was identified by the neutral loss. The intensity of each sterol was normalized to the total lipid signal, which was reported as a percentage of all lipids.
The absolute levels of free cholesterol and cholesteryl esters in the liver were determined by first homogenizing 20 mg of frozen liver in 400 μl of chloroform/isopropyl alcohol/Nonidet P-40 (7:11:0.1). The homogenates were then centrifuged at 15,000 × g for 10 min at 4 °C. The supernatant from this spin was subjected to quantification of free cholesterol and cholesterol ester using a cholesterol/cholesteryl ester quantitation kit (Abcam, Cambridge, UK) according to the manufacturer's procedure.
Cholesterol and Fatty Acid Synthesis in Vivo
Rates of cholesterol and fatty acid synthesis were measured in control WT and hmgcr knock-in mice fed ad libitum with chow diet using 3H-labeled water as described previously (27). The rates of cholesterol and fatty acid synthesis were calculated as nmol of 3H-labeled water incorporated into fatty acids or digitonin-precipitable sterols/h/g of tissue.
Results
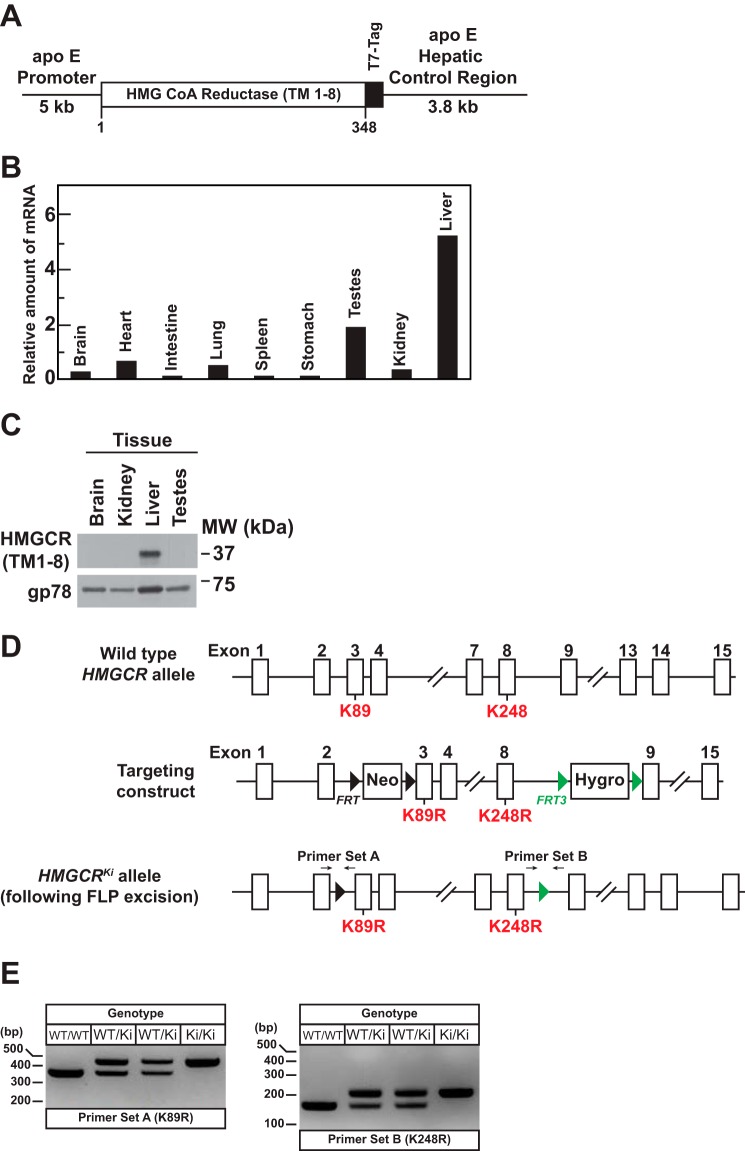
Fig. 1A shows a schematic of a transgene encoding transmembrane domains 1–8 of hamster HMGCR (HMGCR (TM1–8)) fused to three T7 epitopes that was used to generate transgenic mice hereafter designated Tg-HMGCR (TM1–8). Liver-directed expression of HMGCR (TM1–8) was driven by the apoE promoter and its hepatic control region (22). Tg-HMGCR (TM1–8) mice were maintained as hemizygotes by breeding with WT C57BL/6 mice. As expected, expression of HMGCR (TM1–8) mRNA and protein was highest in livers of transgenic mice (Fig. 1, B and C). Tg-HMGCR (TM1–8) mice were grossly indistinguishable from WT littermates normal and had similar body and liver weights as their WT counterparts. No significant differences were observed in plasma and hepatic levels of cholesterol, triglycerides, and free fatty acids between WT and transgenic mice (data not shown).
FIGURE 1.
Generation of Tg-HMGCR (TM1–8) mice expressing HMGCR (TM1–8) in the liver and hmgcr knock-in mice expressing ubiquitination-resistant HMGCR. A, schematic of transgenic construct used to generate Tg-HMGCR (TM1–8) mice. The transgenic construct contains a cDNA fragment encoding transmembrane domains 1–8 (corresponding to amino acids 1–348) of hamster HMG-CoA reductase followed by three copies of the T7 epitope under control of the human apoE promoter and its hepatic control region. B, total RNA extracted from the indicated tissues of four male Tg-HMGCR (TM1–8) mice (12–14 weeks of age) fed ad libitum a chow diet was pooled and subjected to quantitative real-time RT-PCR using transgene-specific primers as described under “Experimental Procedures.” The relative amount of transgene mRNA was calculated using the comparative threshold cycle (CT) method and the mouse glyceraldehyde 3-phosphate dehydrogenase mRNA as an invariant control. C, detergent lysates of the indicated tissue from the same animals used in B were prepared and pooled as described under “Experimental Procedures.” Aliquots of pooled lysates (45 μg of protein/lane) were subjected to SDS-PAGE, and immunoblot analysis was carried out with anti-T7 IgG (against HMGCR (TM1–8)) and anti-gp78 IgG. D, targeting strategy for constructing the hmgcr knock-in allele harboring mutations of lysines 89 (K89R) and 248 (K248R) to arginine. Flippase recognition target (FRT) sites and FRT3 sites are indicated by small black and green triangles, respectively. Neo, neomycin resistance gene; Hygro, hygromycin resistance gene. The location of two primer sets used for genotyping is denoted by arrows. E, genomic DNA isolated from the tails of mice of the indicated genotype were amplified by PCR using primer set A and primer set B and fractionated on 2% agarose gels. Bands corresponding to the K89R and K248R alleles were visualized by staining of gels with ethidium bromide.
Fig. 1D shows the strategy utilized to generate hmgcr knock-in mice (hmgcrKi/Ki) in which lysine residues 89 and 248 were replaced with arginines. For all experiments described here, hmgcrWT/Ki heterozygous male and female mice were crossed to obtain WT and hmgcrKi/Ki littermates (Fig. 1E). Mice homozygous for both knock-in mutations were born at expected Mendelian ratios. WT and hmgcr knock-in littermates were externally indistinguishable and had similar body and liver weights; we also observed no significant differences between WT and hmgcr knock-in mice in plasma cholesterol, triglycerides, and free fatty acids or hepatic triglycerides (data not shown). However, levels of free cholesterol were slightly but significantly increased in hmgcr knock-in mouse livers, which was accompanied by a 2–3-fold increase in hepatic cholesteryl esters (Fig. 2 (A and B) and Table 1).
TABLE 1.
Comparison of hepatic neutral lipid content between WT and hmgcr knock-in mice
Male mice (7–8 weeks old) were fed a chow diet ad libitum before study. WT mice were littermates of hmgcrKi/Ki mice. Each value represents the mean ± S.E. of 8–9 values.
| Lipid class | Percentage of total lipids | -Fold changea | p | No. observed | No. changed |
|---|---|---|---|---|---|
| % | -fold | ||||
| Neutral lipid | 61.5279 | 0.911 | 0.162 | 516 | 170 |
| Triacylglycerols | 61.3417 | 0.908 | 0.154 | 482 | 151 |
| Diacylglycerols | 0.1236 | 1.471 | 0.035 | 26 | 11 |
| Cholesteryl esters | 0.0625 | 2.704 | 0.004 | 8 | 8 |
a -Fold changes are expressed as mean intensities obtained for knock-in mice divided by those obtained for WT counterparts.
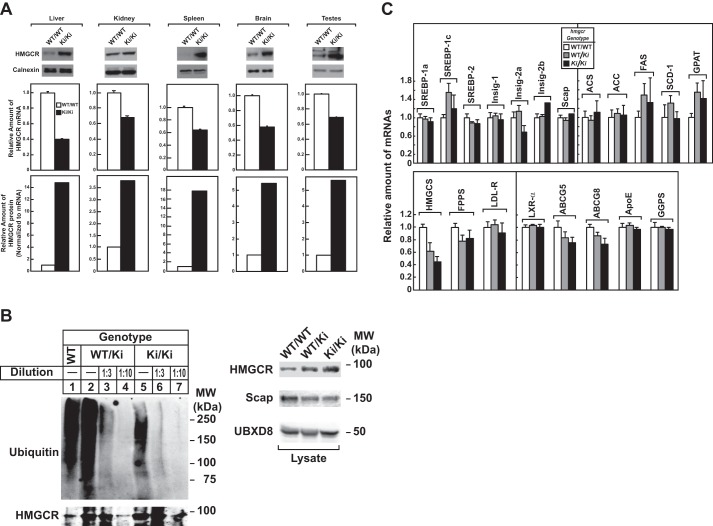
Immunoblot analysis revealed that livers of hmgcrWT/Ki and hmgcrKi/Ki mice fed a chow diet ad libitum exhibited a marked increase in HMGCR protein as compared with that in WT littermates, despite a reduction in HMGCR mRNA (Fig. 3A). When normalized to the amount of hepatic HMGCR mRNA, we estimate that HMGCR protein accumulated ∼6- and 14-fold, respectively, in livers of hmgcrWT/Ki and hmgcrKi/Ki mice relative to WT animals. The relative amount of HMGCR protein (normalized to tissue HMGCR mRNA) was increased between 3- and 18-fold in other tissues of hmgcrKi/Ki mice, including the kidney, spleen, brain, and testes (Fig. 4A). These results indicate that mutation of lysine residues 89 and 248 to arginine blocks ubiquitination and subsequent degradation of hepatic HMGCR, causing the enzyme to accumulate. Indeed, Fig. 4B reveals that ubiquitination of HMGCR protein was reduced despite its accumulation in livers of hmgcrWT/Ki and hmgcrKi/Ki mice (compare lanes 1, 2, and 5). Similar results were observed in an independent repeat experiment (data not shown). Notably, ubiquitination of HMGCR in knock-in mice was not completely eliminated, which could result from ubiquitination at the protein's N terminus (28).
FIGURE 4.
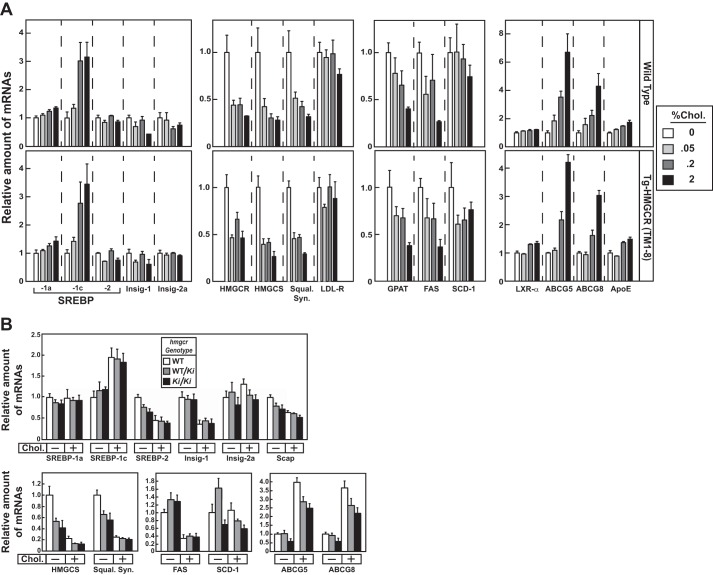
Hepatic HMGCR accumulates in tissues of hmgcr knock-in mice, due to resistance to ubiquitination. A, membrane extract fractions were obtained from the liver, kidney, spleen, brain, and testis of 6–7-week-old male WT and hmgcrKi/Ki mice fed an ad libitum chow diet (5 mice/group). Aliquots of membrane extract fractions for each group were pooled and subjected to immunoblotting (30 μg/lane) for HMGCR and calnexin (top). Total RNA from each tissue was reverse-transcribed, and aliquots of cDNA were pooled for each group. cDNA was subjected to quantitative real-time PCR (middle) as indicated under “Experimental Procedures.” The relative amount of HMGCR protein was obtained as described in the legend to Fig. 3 (bottom). Each bar represents the mean ± S.E. (error bars) of triplicate samples. B, 8–10-week-old male WT, hmgcrWT/Ki, and hmgcrKi/Ki littermates (4 mice/group) were fed an ad libitum chow diet before study. Aliquots of liver lysates for each group were pooled and immunoprecipitated with anti-HMGCR polyclonal antibodies and immunoblotted for ubiquitin or HMGCR (left). To adjust the amount of HMGCR protein subjected to immunoprecipitation, liver lysates were diluted as indicated. Ten percent of the lysates were subjected to immunoblotting for HMGCR, Scap, and UBXD8 (right). C, total RNA from livers of mice used in Fig. 3 was separately isolated. Equal amounts of RNA from individual mice were subjected to quantitative real-time PCR using apoB mRNA as an invariant control. Each value represents the amount of mRNA relative to that in WT mice, which was arbitrarily set as 1. Each bar represents the mean ± S.E. of data from five mice. FPPS, farnesyl pyrophosphate synthase; HMGCS, HMG coenzyme A synthase; LDL-R, LDL-receptor; FAS, fatty acid synthase; SCD-1, stearoyl coenzyme A desaturase-1; GGPS, geranylgeranyl pyrophosphate synthase; GPAT, glycerol-3-phosphate acyltransferase; ACS, acetyl coenzyme A synthetase; ACC, acetyl coenzyme A carboxylase; ABCG5 and ABCG8, ATP-binding cassette subfamily G member 5 and 8, respectively.
Comparison of various components of the Scap-SREBP pathway in WT and hmgcr knock-in mice shows that precursor and nuclear forms of SREBP-2 were reduced in livers of hmgcrWT/Ki and hmgcrKi/Ki mice (Fig. 3B). This reduction was associated with elevated sterol levels in livers of hmgcr knock-in mice (Fig. 2 (A–C) and Tables 1–3) and reduced levels (20–70%) of mRNAs encoding HMGCR and the other SREBP-2 target genes encoding cholesterol biosynthetic enzymes (Fig. 4C and data not shown). Hepatic sterol synthesis was similar between WT and hmgcr knock-in mice (Fig. 2D); however, the reaction was significantly enhanced in testes and brains of hmgcr knock-in mice despite a reduction in SREBP-2 target genes in the tissues (data not shown). In addition, levels of cholesteryl esters were increased in testes and brains of knock-in mice, but the difference observed in brains was not statistically significant (Fig. 2E).
TABLE 2.
Comparison of biliary sterol content between WT and hmgcr knock-in mice
Male mice (7–8 weeks old) were fed a chow diet ad libitum before study. WT mice were littermates of hmgcrKi/Ki mice. Each value represents the mean ± S.E. of 8–9 values.
| Sterol | Wild type | Knock-in | p |
|---|---|---|---|
| mg/dl | mg/dl | ||
| Lanosterol | 0.63 ± 0.13 | 1.21 ± 0.14 | 0.009 |
| Lathosterol | 0.16 ± 0.03 | 0.29 ± 0.04 | 0.015 |
| Desmosterol | 0.58 ± 0.05 | 1.05 ± 0.14 | 0.006 |
| Cholesterol | 116.39 ± 13.10 | 147.98 ± 14.20 | 0.124 |
| Camptosterol | 7.44 ± 0.75 | 7.56 ± 0.87 | 0.922 |
| Sitosterol | 3.01 ± 0.38 | 2.87 ± 0.24 | 0.752 |
TABLE 3.
Comparison of fecal sterol content between WT and hmgcr knock-in mice
Male mice (7–8 weeks old) were fed a chow diet ad libitum before study. WT mice were littermates of hmgcrKi/Ki mice. Each value represents the mean ± S.E. of 8–9 values.
| Sterol | Wild type | Knock-in | p |
|---|---|---|---|
| mg/g feces | mg/g feces | ||
| Lanosterol | 114.56 ± 7.17 | 156.18 ± 6.76 | 0.001 |
| Lathosterol | 143.30 ± 24.82 | 216.47 ± 43.75 | 0.180 |
| Desmosterol | 34.01 ± 4.57 | 39.07 ± 3.95 | 0.412 |
| Cholesterol | 887.95 ± 67.74 | 1027.45 ± 60.17 | 0.143 |
| Camptosterol | 813.72 ± 21.94 | 855.08 ± 25.38 | 0.242 |
| Sitosterol | 2179.20 ± 72.10 | 2243.83 ± 137.54 | 0.694 |
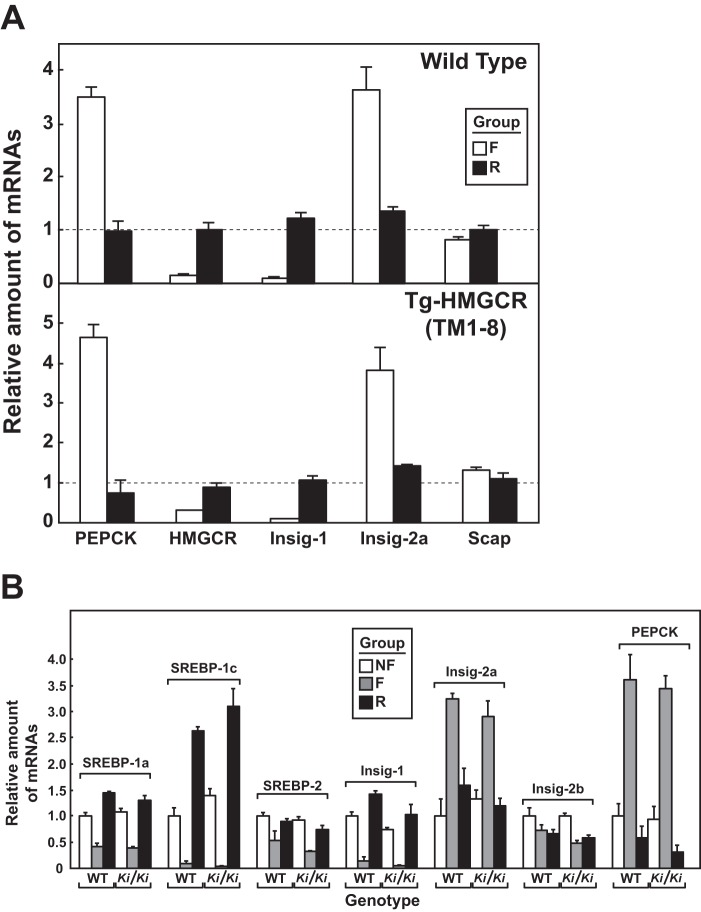
In contrast to SREBP-2, nuclear SREBP-1 was increased in livers of hmgcr knock-in mice (Fig. 3B). This increase probably results from sterol-mediated activation of liver X receptors (LXRs) that modulate expression of SREBP-1c, the major SREBP-1 isoform expressed in mouse livers (29). Indeed, mRNAs encoding SREBP-1c and its target genes, including fatty acid synthase, stearoyl coenzyme A desaturase-1, and glycerol-3-phosphate acyltransferase, were elevated in livers of hmgcr knock-in mice (Fig. 4C).
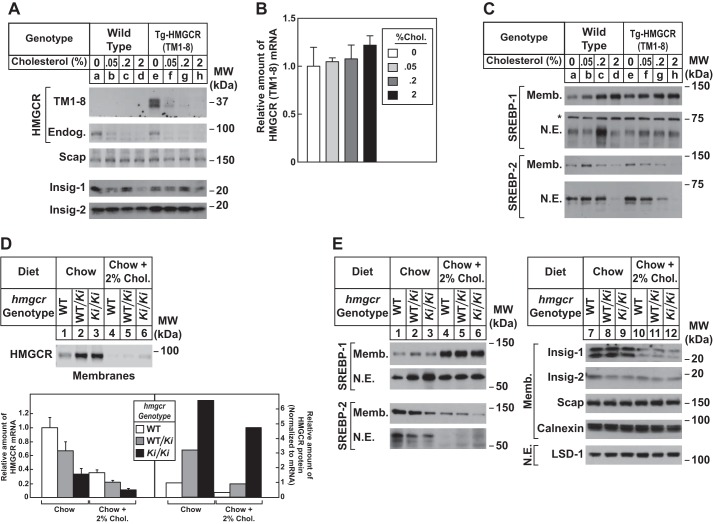
Fig. 5A compares expression of HMGCR (TM1–8) protein in livers of Tg-HMGCR (TM1–8) mice fed diets containing varying amounts of cholesterol. When transgenic mice were fed as little as 0.05% cholesterol, the level of HMGCR (TM1–8) protein was substantially reduced in hepatic membranes (Fig. 5A, compare lanes e and f); higher amounts of cholesterol led to the complete disappearance of HMGCR (TM1–8) (lanes g and h). Cholesterol feeding failed to inhibit expression of HMGCR (TM1–8) mRNA (Fig. 5B).
FIGURE 5.
Dietary cholesterol suppresses expression of HMGCR (TM1–8) in Tg-HMGCR (TM1–8) mouse livers and endogenous HMGCR in hmgcr knock-in mouse livers. A–C, male mice (6–8 weeks of age, 4 mice/group) were fed an ad libitum chow diet supplemented with the indicated amount of cholesterol for 5 days. A and C, aliquots of membrane (Memb.) and nuclear extract (N.E.) fractions from homogenized livers (10–30 μg of total protein/lane) were analyzed by immunoblot analysis with anti-T7 IgG (against HMGCR (TM1–8)) and antibodies against the indicated proteins. B, equal amounts of RNA from the individual mice used in A and C were subjected to quantitative real-time RT-PCR using primers against the HMGCR (TM1–8) mRNA; apoB mRNA was used as an invariant control. Values represent the amount of HMGCR (TM1–8) mRNA relative to that in transgenic mice fed a chow diet, which is arbitrarily defined as 1. Bars, mean ± S.E. (error bars) of data from four mice. *, a nonspecific cross-reactive band. D and E, male WT, hmgcrWT/Ki, and hmgcrKi/Ki littermates (6–8 weeks of age, 4 mice/group) were fed an ad libitum chow diet supplemented with 2% cholesterol as indicated for 5 days. Aliquots of membrane (30 μg of protein/lane) and nuclear extract (20–50 μg of protein/lane) fractions from homogenized livers were analyzed by immunoblot as described in the legend to Fig. 3. Although shown in separate panels, Scap and calnexin serve as loading controls for the HMGCR immunoblot. For mRNA analysis (D), equal amounts of RNA from individual mice were subjected to quantitative real-time RT-PCR as described in the legend to Fig. 3A. Values represent the amount of mRNA relative to that in WT mice, which was arbitrarily set as 1. Bars, mean ± S.E. of data from four mice. The relative amount of HMGCR protein in hmgcr knock-in mice was determined as described in the legend to Fig. 3.
Dietary cholesterol caused a reduction of endogenous HMGCR protein in membranes from livers of WT (Fig. 5A, lanes a–d) and Tg-HMGCR (TM1–8) mice (lanes e–h). Membrane-bound precursor and nuclear forms of SREBP-2 were also reduced in livers of cholesterol-fed animals (Fig. 5C). Levels of SREBP-1 precursor (Fig. 5C) and mRNA (Fig. 6A) were increased in livers of WT and transgenic mice fed cholesterol, whereas nuclear SREBP-1 levels remained largely unchanged. This is probably due to combined effects of cholesterol on ER to Golgi transport of Scap-SREBP and activation of LXRs (30). As expected, expression of mRNAs encoding SREBP-2 target genes was reduced to similar levels in cholesterol-fed WT and Tg-HMGCR (TM1–8) mice (Fig. 6A).
FIGURE 6.
Effect of dietary cholesterol on expression of mRNAs encoding components of the Scap-SREBP pathway in livers of Tg-HMGCR (TM1–8) and hmgcr knock-in mice. Total RNA from livers of mice used in Fig. 5, A and D (4 mice/group) was separately isolated. Equal amounts of RNA from the individual mice were subjected to quantitative real-time RT-PCR using primers against the indicated gene; apoB mRNA was used as an invariant control. Each value represents the amount of mRNA relative to that in WT and transgenic mice (A) or WT mice (B) fed a chow diet, which is arbitrarily defined as 1. Bars, mean ± S.E. (error bars) of data from four mice. Squal. Syn., squalene synthase.
Fig. 5D shows that elevated levels of HMGCR protein in hmgcrWT/Ki and hmgcrKi/Ki mice were reduced by cholesterol feeding to an extent similar to that observed in WT mice (compare lanes 1-3 with lanes 4-6) (Table 4). Dietary cholesterol also reduced the mRNA for HMGCR, Insig-1, and other SREBP target genes in WT and hmgcr knock-in mice (Figs. 5D and 6B), which resulted from inhibition of SREBP-2 processing (Fig. 5E, compare lanes 1-3 with lanes 4-6). Notably, Insig-1 protein levels were also reduced upon cholesterol feeding (Fig. 5E, lanes 7–12). Levels of the SREBP-1 precursor were elevated by cholesterol feeding in WT and knock-in mice (Fig. 5E, compare lanes 1–3 with lanes 4-6), which correlated with an increase in mRNAs for SREBP-1c and two other LXR target genes, Abcg5 and Abcg8 (Fig. 6B). Processing of SREBP-1 was mostly resistant to dietary cholesterol in WT and hmgcr knock-in animals (Fig. 5E, lanes 4–6).
TABLE 4.
Comparison of WT and hmgcr knock-in mice subjected to cholesterol feeding
8–10-week-old male mice (4/group) were fed a chow diet in the absence or presence of 2% cholesterol for 5 days before study. WT mice were littermates of hmgcrWT/Ki and hmgcrKi/Ki mice. Each value represents the mean ± S.E. of 4 values.
| Parameter | WT (chow) | WT (2% cholesterol) | hmgcrWT/Ki (chow) | hmgcrWT/Ki (2% cholesterol) | hmgcrKi/Ki (chow) | hmgcrKi/Ki (2% cholesterol) |
|---|---|---|---|---|---|---|
| Body weight (g) | 25.9 ± 0.5 | 24.8 ± 1.0 | 25.2 ± 0.2 | 25.3 ± 0.6 | 25.2 ± 0.5 | 25.7 ± 0.8 |
| Liver weight (g) | 1.38 ± 0.20 | 1.56 ± 0.10 | 1.32 ± 0.07 | 1.48 ± 0.03 | 1.41 ± 0.50 | 1.55 ± 0.06 |
| Plasma triglycerides (mg/dl) | 127 ± 4 | 175 ± 13 | 173 ± 14 | 183 ± 7 | 142 ± 24 | 163 ± 11 |
| Plasma cholesterol (mg/dl) | 104 ± 4 | 193 ± 3 | 128 ± 14 | 178 ± 8 | 116 ± 9 | 181 ± 6 |
| Plasma fatty acids (meq/liter) | 1.11 ± 0.06 | 1.24 ± 0.08 | 1.24 ± 0.08 | 1.31 ± 0.06 | 1.00 ± 0.07 | 1.33 ± 0.10 |
| Liver triglycerides (mg/g) | 3.4 ± 0.6 | 25.2 ± 6.5 | 3.3 ± 0.4 | 22.1 ± 5.1 | 7.4 ± 3.1 | 22.2 ± 4.2 |
| Liver cholesterol (mg/g) | 2.1 ± 0.1 | 9.7 ± 0.7 | 2.1 ± 0.1 | 10.5 ± 1.6 | 2.3 ± 0.1 | 11.2 ± 1.2 |
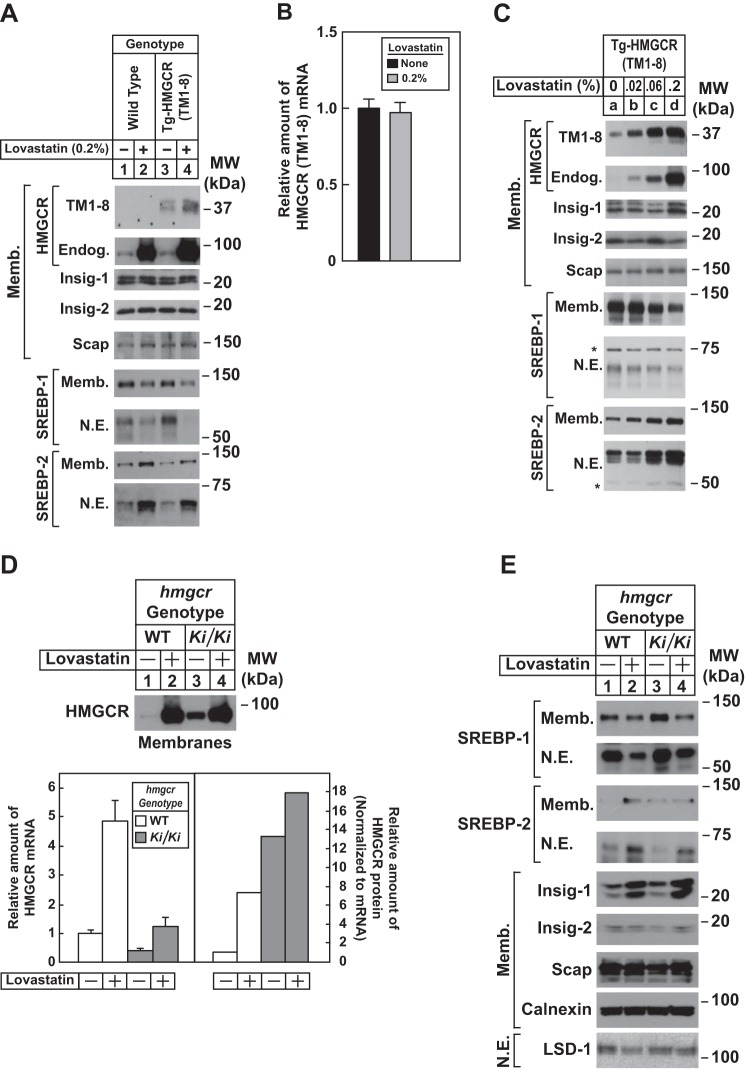
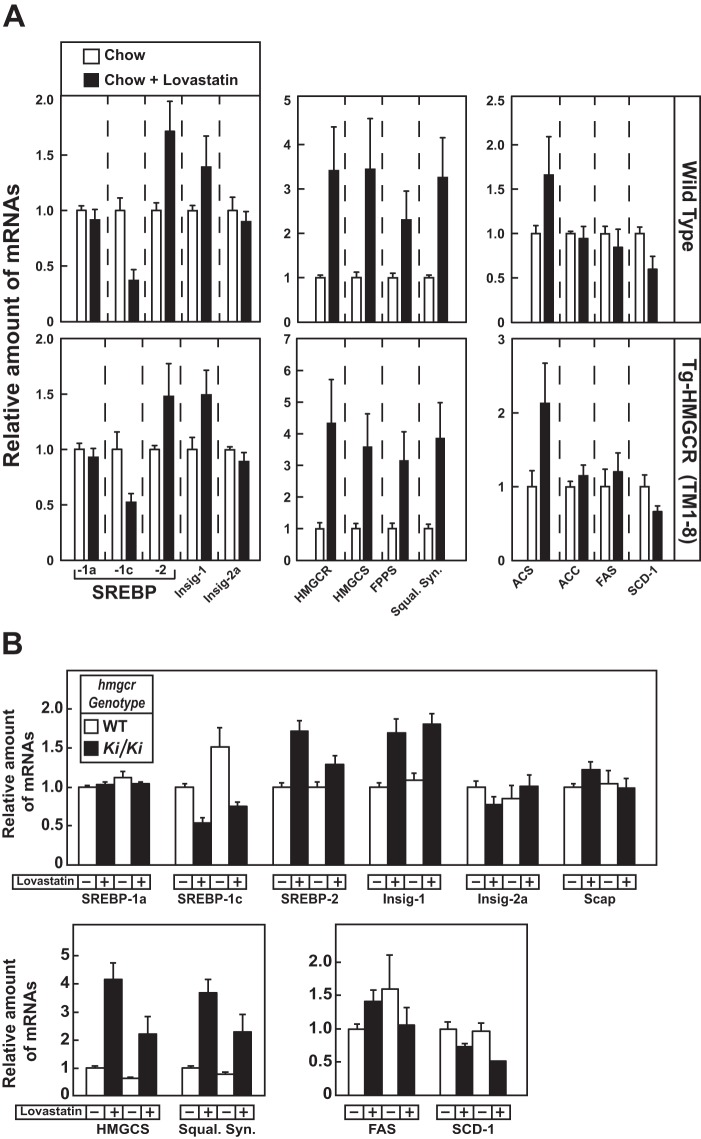
The response of HMGCR (TM1–8) degradation to cholesterol deprivation was examined by feeding Tg-HMGCR (TM1–8) mice a chow diet containing the HMGCR inhibitor lovastatin. Consumption of a chow diet containing 0.2% lovastatin caused an increase in the amount of HMGCR (TM1–8) protein in membranes of transgenic mouse livers (Fig. 7A, compare lanes 3 and 4). However, the treatment did not alter expression of HMGCR (TM1–8) mRNA (Fig. 7B). Endogenous HMGCR protein and mRNA were increased in livers of WT and Tg-HMGCR (TM1–8) mice fed lovastatin (Figs. 7A (compare lanes 1 and 3 with lanes 2 and 4) and 8A). This was associated with an increase in the nuclear content of SREBP-2 in livers of lovastatin-fed WT and transgenic mice (Fig. 7A, lanes 2 and 4) and enhanced expression of mRNAs encoding its target genes (Fig. 8A). In contrast, lovastatin lowered the amount of both precursor and nuclear forms of SREBP-1 (Fig. 7A, compare lanes 1 and 3 with lanes 2 and 4), which can be attributed to loss of an endogenous sterol ligand for LXR (31). In the second cholesterol deprivation experiment, we treated Tg-HMGCR (TM1–8) mice with various amounts of lovastatin. As little as 0.02% lovastatin noticeably stabilized HMGCR (TM1–8) (Fig. 7C, compare lanes a and b); the protein was further stabilized by higher concentrations of the inhibitor (lanes c and d). Precursor and nuclear forms of SREBP-2 and SREBP-1 were induced and suppressed, respectively, by lovastatin in a dose-dependent fashion (Fig. 7C, lanes a–d).
FIGURE 7.
Cholesterol deprivation enhances expression of HMGCR (TM1–8) in livers of transgenic mice and endogenous HMGCR in livers of hmgcr knock-in mice. A–C, male mice (6–8 weeks of age, 5 mice/group in A and B or 3 mice/group in C) were fed an ad libitum chow diet in the absence or presence of the indicated concentration of lovastatin for 5 days. A and C, aliquots of membrane and nuclear extract fractions (10–30 μg protein/lane) from homogenized livers were analyzed by immunoblot as described in the legend to Fig. 5, A and C. Asterisks denote nonspecific cross-reactive bands. Equal amounts of RNA from individual mice used in A were subjected to quantitative real-time RT-PCR as described in the legend to Fig. 5B. Values represent the amount of HMGCR (TM1–8) mRNA relative to that in chow-fed transgenic mice, which is arbitrarily defined as 1. Bars, mean ± S.E. (error bars) of data from five mice. D and E, male WT and hmgcrKi/Ki littermates (6–8 weeks of age, 4 mice/group) were fed an ad libitum chow diet in the absence or presence of 0.2% lovastatin for 5 days. Aliquots of membrane (Memb., 30 μg protein/lane) and nuclear extract (N.E., 20–50 μg of protein/lane) fractions from homogenized livers were analyzed by immunoblot as described in the legend to Fig. 3. Although shown in separate panels, Scap and calnexin serve as loading controls for the HMGCR immunoblot. For mRNA analysis (D), equal amounts of RNA from individual mice were subjected to quantitative real-time RT-PCR as described in the legend to Fig. 3A. Values represent the amount of mRNA relative to that in WT mice, which was arbitrarily set as 1. Bars, mean ± S.E. of data from four mice. The relative amount of HMGCR protein in hmgcr knock-in mice was determined as described in the legend to Fig. 3.
FIGURE 8.
Effect of cholesterol deprivation on expression of mRNAs encoding components of the Scap-SREBP pathway in livers of Tg-HMGCR (TM1–8) and hmgcr knock-in mice. Total RNA from livers of mice used in Fig. 7, A and D (4 or 5 mice/group) was separately isolated. Equal amounts of RNA from the individual mice were subjected to quantitative real-time RT-PCR using primers against the indicated gene; apoB mRNA was used as an invariant control. Each value represents the amount of mRNA relative to that in WT and transgenic mice (A) or WT mice (B) fed a chow diet, which is arbitrarily defined as 1. Bars, mean ± S.E. (error bars) of data from four or five mice.
Fig. 7D shows that lovastatin triggered an increase in HMGCR protein in livers of hmgcr knock-in mice (lanes 3 and 4; see Table 5); however, the increase was blunted compared with that observed in WT mice (lanes 1 and 2; see quantification). This is probably due to accumulation of HMGCR protein combined with increased sterol levels and inhibition of SREBP-2 processing in livers of knock-in mice. Lovastatin inhibited levels of precursor and nuclear forms of SREBP-1 (Fig. 7E, lanes 1–4) and enhanced levels of precursor and nuclear SREBP-2 (lanes 1–4). Furthermore, mRNAs for SREBP-2 and its target genes were elevated upon lovastatin feeding, whereas SREBP-1c mRNA was reduced (Fig. 8B).
TABLE 5.
Comparison of WT and hmgcr knock-in mice subjected to lovastatin feeding
6–7-week-old male mice (4/group) were fed a chow diet in the absence or presence of 0.2% lovastatin for 5 days before study. WT mice were littermates of hmgcrWT/Ki and hmgcrKi/Ki mice. Each value represents the mean ± S.E. of four values.
| Parameter | WT (chow) | WT (0.2% lovastatin) | hmgcrKi/Ki (chow) | hmgcrKi/Ki (0.2% lovastatin) |
|---|---|---|---|---|
| Body weight (g) | 20.2 ± 0.7 | 20.5 ± 0.8 | 20.4 ± 1.1 | 20.1 ± 0.3 |
| Liver weight (g) | 1.16 ± 0.01 | 1.19 ± 0.04 | 1.33 ± 0.04 | 1.11 ± 0.03 |
| Plasma triglycerides (mg/dl) | 135 ± 18 | 142 ± 11 | 140 ± 8 | 209 ± 34 |
| Plasma cholesterol (mg/dl) | 99 ± 3 | 71 ± 5 | 115 ± 6 | 84 ± 12 |
| Plasma fatty acids (meq/liter) | 1.04 ± 0.02 | 1.10 ± 0.14 | 1.08 ± 0.09 | 1.75 ± 0.61 |
| Liver triglycerides (mg/g) | 5.6 ± 1.1 | 4.6 ± 0.4 | 7.1 ± 1.1 | 5.5 ± 0.7 |
| Liver cholesterol (mg/g) | 2.0 ± 0.2 | 2.0 ± 0.2 | 2.2 ± 0.1 | 2.2 ± 0.2 |
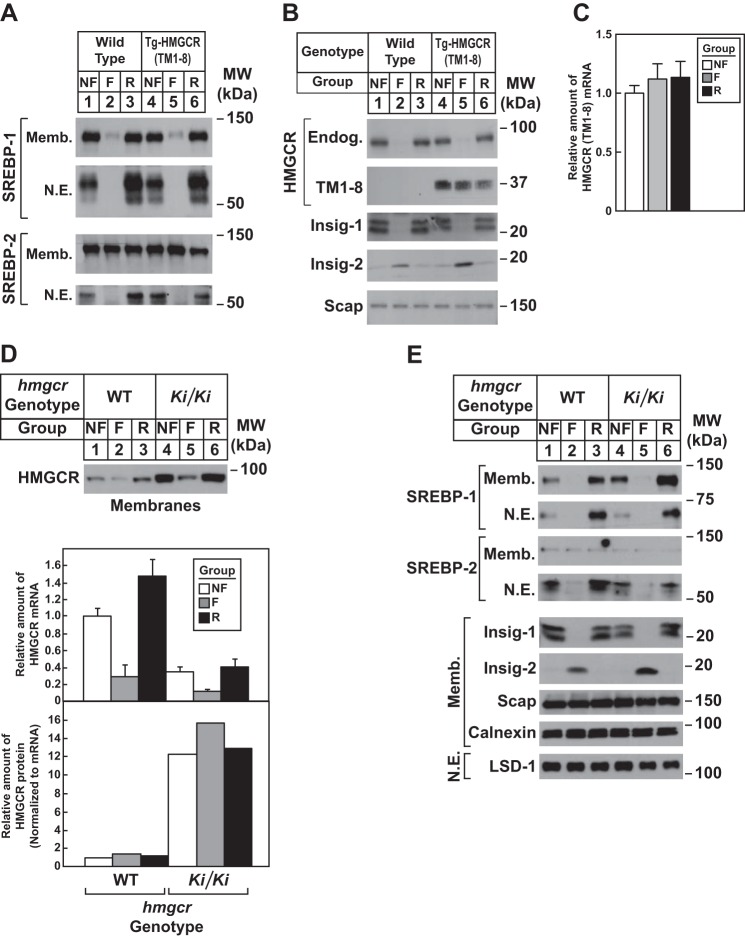
Studies were next carried out to determine whether modulation of Insig levels affected degradation of HMGCR (TM1–8). The hormone insulin reciprocally regulates levels of Insig-1 and Insig-2 in livers of mice (32). To modulate insulin levels in WT and Tg-HMGCR (TM1–8) mice, we subjected the animals to a 12-h fast, which lowered plasma insulin by 80–90%, followed by a 12-h period of refeeding, which increased insulin >10-fold over the fasted level (Table 6). Compared with nonfasted animals, fasting reduced the amount of precursor and nuclear forms of SREBP-1 in WT and transgenic mice (Fig. 9A, lanes 1, 2, 4, and 5 refeeding restored levels of SREBP-1 precursor and caused an overshoot in the nuclear form of the protein (lanes 3 and 6). Fasting also reduced nuclear SREBP-2, and refeeding restored these levels (Fig. 9A, lanes 2, 3, 5, and 6). As reported previously, fasting caused the disappearance of Insig-1 (due to the down-regulation of nuclear SREBP) and the appearance of Insig-2 (Fig. 9B, compare lanes 1 and 4 with lanes 2 and 5). These changes in Insig-1 and Insig-2 protein levels were completely reversed by refeeding (lanes 3 and 6). Endogenous HMGCR protein was reduced upon fasting of WT and transgenic mice (Fig. 9B, lanes 2 and 5); its expression was restored by refeeding (lanes 3 and 6). The mRNAs for HMGCR, Insig-1, and Insig-2a mRNAs varied with fasting/refeeding in a manner mirroring that of their respective proteins (Fig. 10A). In contrast to endogenous HMGCR, levels of HMGCR (TM1–8) protein (Fig. 9B, lanes 4-6) and mRNA (Fig. 9C) were not reduced in livers of fasted transgenic mice.
TABLE 6.
Comparison of WT and Tg-HMGCR (TM1–8) mice subjected to fasting and refeeding
Male mice (7–8 weeks of age) were subjected to fasting and refeeding as described under “Experimental Procedures.” WT mice were littermates of transgenic mice. Each value represents the mean ± S.E. of five mice.
| WT |
Tg-HMGCR (TM1–8) |
|||||
|---|---|---|---|---|---|---|
| Nonfasted | Fasted | Refed | Nonfasted | Fasted | Refed | |
| Body weight, pretreatment (g) | 25.0 ± 0.8 | 24.9 ± 1.2 | 24.9 ± 0.7 | 25.0 ± 0.5 | 24.8 ± 0.6 | 26.6 ± 0.4 |
| Body weight, post-treatment (g) | 24.9 ± 0.6 | 22.7 ± 1.0 | 24.8 ± 0.8 | 26.1 ± 0.4 | 22.3 ± 0.8 | 26.9 ± 0.5 |
| Liver weight (g) | 1.25 ± 0.13 | 1.11 ± 0.09 | 1.48 ± 0.10 | 1.41 ± 0.03 | 0.98 ± 0.06 | 1.75 ± 0.03 |
| Liver cholesterol (mg/g) | 2.7 ± 0.1 | 2.8 ± 0.1 | 2.1 ± 0.0 | 2.3 ± 0.1 | 3.4 ± 0.0 | 2.2 ± 0.2 |
| Liver triglyceride (mg/g) | 6.4 ± 1.1 | 39.1 ± 8.1 | 9.0 ± 1.8 | 6.8 ± 1.1 | 44.0 ± 4.9 | 9.1 ± 1.5 |
| Plasma fatty acids (meq/liter) | 0.49 ± 0.03 | 0.99 ± 0.16 | 0.41 ± 0.07 | 0.48 ± 0.11 | 0.90 ± 0.12 | 0.28 ± 0.02 |
| Plasma triglycerides (mg/dl) | 236 ± 29 | 230 ± 20 | 263 ± 33 | 263 ± 12 | 219 ± 11 | 334 ± 15 |
| Plasma cholesterol (mg/dl) | 113 ± 10 | 126 ± 6 | 109 ± 2 | 118 ± 11 | 127 ± 2 | 118 ± 3 |
| Plasma glucose (mg/dl) | 152 ± 3 | 67 ± 9 | 133 ± 6 | 157 ± 5 | 69 ± 0 | 119 ± 4 |
| Plasma insulin (ng/ml) | 2.35 ± 0.15 | 0.24 ± 0.04 | 2.74 ± 0.96 | 1.39 ± 0.12 | 0.24 ± 0.02 | 5.48 ± 0.48 |
FIGURE 9.
Effect of fasting and refeeding on expression of HMGCR (TM1–8) in livers of transgenic mice and endogenous HMGCR in livers of hmgcr knock-in mice. A–C, male WT and Tg-HMGCR (TM1–8) mice (6–8 weeks of age, 4 mice/group) were subjected to fasting and refeeding as described under “Experimental Procedures.” A and B, aliquots of membrane and nuclear extract fractions from homogenized livers (10–30 μg of protein/lane) were analyzed by immunoblot as described in the legend to Fig. 5. C, equal amounts of RNA from individual mice used in A and B were subjected to quantitative real-time RT-PCR as described in the legend to Fig. 5B. Values represent the amount of HMGCR (TM1–8) mRNA relative to that in chow-fed transgenic mice, which is arbitrarily defined as 1. Bars, mean ± S.E. (error bars) of data from four mice. Metabolic parameters of WT and Tg-HMGCR (TM1–8) mice subjected to fasting and refeeding are provided in Table 4. D and E, male WT and hmgcrKi/Ki littermates (6–8 weeks of age, 4 mice/group) were subjected to fasting and refeeding as described in A. Aliquots of membrane (Memb., 30 μg of protein/lane) and nuclear extract (N.E., 20–50 μg of protein/lane) fractions from homogenized livers were subjected to immunoblot analysis as described in the legend to Fig. 3. Although shown in separate panels, Scap and calnexin serve as loading controls for the HMGCR immunoblot. Equal amounts of RNA from individual mice were subjected to quantitative real-time RT-PCR as described in the legend to Fig. 3A. Values represent the amount of mRNA relative to that in WT nonfasted mice, which was arbitrarily set as 1. Bars, mean ± S.E. of data from four mice. The relative amount of HMGCR protein in hmgcr knock-in mice was determined as described in the legend to Fig. 3.
FIGURE 10.
Effect of fasting and refeeding on expression of mRNAs encoding components of the Scap-SREBP pathway in livers of Tg-HMGCR (TM1–8) and hmgcr knock-in mice. Total RNA from livers of mice used in Fig. 9, A and D (4 mice/group) was separately isolated. Equal amounts of RNA from individual mice were subjected to quantitative real-time RT-PCR using primers against the indicated gene; apoB mRNA was used as an invariant control. Each value represents the amount of mRNA relative to that in control, nonfasted mice, which was arbitrarily set as 1. Bars, mean ± S.E. (error bars) of data from four mice. PEPCK, phosphoenolpyruvate carboxykinase.
To confirm that fasting reduced levels of endogenous HMGCR through inhibition of SREBP-mediated transcription rather than sterol-accelerated degradation, we conducted the experiment shown in Fig. 9D (see Table 7). The results show that fasting reduced levels of HMGCR protein and mRNA in both WT and hmgcr knock-in mice (Fig. 9D, lanes 2 and 5). However, when normalized to the amount of HMGCR mRNA, the relative amount of HMGCR protein was unchanged in livers of fasted mice. Precursor and nuclear forms of SREBP-1 and SREBP-2 were reduced by fasting (Fig. 9E, lanes 2 and 5) and restored by refeeding (lanes 3 and 6). Insig-1 and -2 were reciprocally regulated at the protein (Fig. 9E, lanes 2, 3, 5, and 6) and mRNA levels (Fig. 10B) by fasting and refeeding.
TABLE 7.
Metabolic parameters of WT and hmgcr knock-in mice subjected to fasting and refeeding
Male mice (7–8 weeks of age) were subjected to fasting and refeeding as described under “Experimental Procedures.” WT mice were littermates of hmgcrWT/Ki and hmgcrKi/Ki mice. Each value represents the mean ± S.E. of four mice.
| WT |
hmgcrKi/Ki |
|||||
|---|---|---|---|---|---|---|
| Nonfasted | Fasted | Refed | Nonfasted | Fasted | Refed | |
| Liver cholesterol (mg/g) | 2.1 ± 0.1 | 3.5 ± 0.2 | 2.2 ± 0.1 | 2.5 ± 0.1 | 3.9 ± 0.1 | 2.5 ± 0.1 |
| Liver triglyceride (mg/g) | 5.2 ± 2.1 | 33.1 ± 6.9 | 6.1 ± 0.6 | 3.7 ± 0.6 | 50.4 ± 3.2 | 5.1 ± 0.8 |
| Plasma fatty acids (meq/liter) | 0.64 ± 0.05 | 1.65 ± 0.30 | 0.40 ± 0.04 | 0.64 ± 0.01 | 1.76 ± 0.21 | 0.43 ± 0.08 |
| Plasma triglycerides (mg/dl) | 182 ± 17 | 279 ± 27 | 206 ± 25 | 186 ± 10 | 211 ± 10 | 172 ± 19 |
| Plasma cholesterol (mg/dl) | 127 ± 13 | 145 ± 11 | 87 ± 6 | 134 ± 12 | 144 ± 9 | 116 ± 9 |
| Plasma glucose (mg/dl) | 113 ± 16 | 45 ± 11 | 83 ± 13 | 102 ± 7 | 44 ± 4 | 75 ± 9 |
| Plasma insulin (ng/ml) | 0.70 ± 0.14 | 0.16 ± 0.03 | 1.82 ± 0.63 | 0.50 ± 0.90 | 0.21 ± 0.05 | 1.01 ± 0.26 |
| Plasma insulin (ng/ml) | 2.35 ± 0.15 | 0.24 ± 0.04 | 2.74 ± 0.96 | 1.39 ± 0.12 | 0.24 ± 0.02 | 5.48 ± 0.48 |
Discussion
Two lines of genetically manipulated mice were used to determine whether sterols modulate degradation of HMGCR in the liver and how sterol-accelerated ERAD contributes to multivalent feedback regulation of cholesterol homeostasis. Characterization of Tg-HMGCR (TM1–8) mice, which express HMGCR (TM1–8) in the liver, revealed that cholesterol feeding caused the protein to disappear from hepatic membranes (Fig. 5A). Conversely, HMGCR (TM1–8) accumulated in hepatic membranes when diets were supplemented with the HMGCR inhibitor lovastatin to deplete cholesterol (Fig. 7, A and C). Importantly, mRNA encoding HMGCR (TM1–8) remained unchanged, regardless of feeding regimen (Figs. 5B and 7B). Together, these results indicate that changes in expression of HMGCR (TM1–8) protein upon cholesterol and lovastatin feeding resulted from sterol-mediated modulation of its degradation.
Knock-in mice expressing ubiquitination-resistant HMGCR accumulated significant amounts of the protein in the liver and other tissues, although levels of HMGCR mRNA were reduced (Figs. 3A and 4A). The decline in HMGCR mRNA can be attributed to reduced levels of nuclear SREBP-2 (Fig. 3B), due to accumulation of cholesterol (Fig. 2). In contrast, levels of nuclear SREBP-1 were elevated in livers of hmgcr knock-in mice, which probably results from activation of LXRs in response to cholesterol accumulation. HMGCR protein also accumulates disproportionately to its mRNA in livers of mice deficient in gp78 (11), a ubiquitin ligase that facilitates HMGCR ubiquitination. However, it should be noted that levels of Insig-2 and, to a lesser extent, Insig-1 protein are increased in gp78-deficient livers. As a result, processing of both SREBP-1 and SREBP-2 is inhibited. Thus, changes in cholesterol metabolism observed in gp78-deficient livers cannot be solely attributed to defects in degradation of HMGCR.
Fig. 2 shows that whereas levels of free and esterified cholesterol were elevated in livers of hmgcr knock-in mice, incorporation of [3H]water into sterols was unchanged. This discrepancy could be explained in part by increases in some of the sterol intermediates, which could contribute to the increase observed in [3H]water studies that measure synthesis of all digitonin-precipitable sterols. It should be noted that accumulation of sterol intermediates has also been observed in Insig-deficient mice and interferes with normal fusion of midline facial structures, producing cleft palate (33). The exact mechanism whereby these sterol intermediates accumulate is unknown and merits further investigation.
Experiments in which cholesterol levels were modulated by diet were conducted to examine the role of sterol-regulated ubiquitination and degradation of HMGCR in regulation of cholesterol homeostasis. When normalized to its cognate mRNA, the relative amount of HMGCR protein was reduced by ∼70% in livers of WT mice subjected to cholesterol feeding (Fig. 5D). However, mRNA-normalized HMGCR protein was reduced by <30% in livers of cholesterol-fed hmgcr knock-in mice, indicating resistance to sterol-accelerated degradation. Despite this resistance, the absolute amount of HMGCR protein in knock-in mouse livers was markedly diminished by cholesterol feeding, which was accompanied by a similar reduction in HMGCR mRNA. Thus, we conclude that feeding mice high levels of cholesterol reduces HMGCR levels primarily through reduced transcription of the HMGCR gene resulting from sterol-mediated inhibition of SREBP-2 activation. Cholesterol depletion studies show that lovastatin-induced accumulation of HMGCR protein was blunted in hmgcr knock-in mice compared with their WT littermates (Fig. 7D). This indicates that inhibition of degradation significantly contributes to the increase in HMGCR protein that occurs upon lovastatin treatment, which depletes not only sterols but also nonsterol isoprenoids that modulate HMGCR degradation (21). Taken together, results of Figs. 5D and 7D reveal that sterol-induced ubiquitination and degradation play a direct and significant role in feedback regulation of HMGCR and cholesterol homeostasis in vivo.
An unexpected result was obtained when Tg-HMGCR (TM1–8) and hmgcr knock-in mice were subjected to a fasting and refeeding regimen. Fasting, which lowers plasma insulin, inhibited activation of SREBPs as indicated by the fall in nuclear SREBP-1 and SREBP-2 as well as mRNA and protein for SREBP targets Insig-1 and HMGCR (Figs. 9 and 10). In contrast to results with endogenous HMGCR, fasting failed to accelerate degradation of HMGCR (TM1–8) (Fig. 9B). This observation is substantiated by results with hmgcr knock-in mice, which show that fasting reduced mRNA encoding endogenous HMGCR but did not accelerate degradation of HMGCR protein (Fig. 9D). Thus, fasting appears to represent a condition under which transcription of the HMGCR gene and degradation of HMGCR protein are uncoupled. This uncoupling could, in part, result from a fasting-induced post-translational modification that prevents incorporation of Scap and its bound SREBP into COPII vesicles destined for the Golgi. Alternatively, the uncoupling of ER to Golgi transport of Scap and accelerated degradation of reductase may result from differential affinity of the two proteins for Insigs whose levels are reciprocally regulated during fasting. It will be important in future studies to appraise these notions and determine the underlying basis for uncoupling of transcriptional and post-transcriptional regulation of HMGCR during fasting.
The importance of the regulatory system that governs feedback regulation of reductase is highlighted by the widespread use of statins to lower plasma levels of LDL-cholesterol and reduce the incidence of atherosclerosis and associated cardiovascular disease (15). However, statins inhibit production of sterol and nonsterol isoprenoids that mediate feedback regulation of reductase. Animals and humans respond to this inhibition by developing high levels of reductase in the liver (34, 35), prompting the need for high levels of the drugs to maintain reductase inhibition and cholesterol lowering. The current analysis of Tg-HMGCR (TM1–8) and hmgcr knock-in mice provides direct evidence that sterol-accelerated degradation significantly contributes to feedback regulation of HMGCR. These novel animal models may prove useful in development of new drugs that accelerate HMGCR degradation, thereby preventing the accumulation of the enzyme associated with statin therapy. These new drugs may improve the effectiveness of statins or provide alternative therapies.
Author Contributions
S. H. and I. Z. H. designed research, performed experiments, analyzed data, and drafted the manuscript; L. N. C., K. G., G. A. Y., M. A. M., J. M., F. X., and L. E. designed the research, performed experiments, and analyzed data; and R. A. D.-B. designed the research, analyzed data, and drafted the manuscript.
Acknowledgments
We thank Guosheng Liang for helpful advice and Norma Anderson for help with in vivo sterol synthesis experiments.
This work was supported by National Institutes of Health Grant HL20948 and an Early Career Scientist Award from the Howard Hughes Medical Institute (to R. D. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HMG
- 3-hydroxy-3-methylglutaryl
- HMGCR
- 3-hydroxy-3-methylglutaryl coenzyme A reductase
- ER
- endoplasmic reticulum
- SREBP
- sterol regulatory element-binding proteins
- LXR
- liver X receptor.
References
- 1. Goldstein J. L., and Brown M. S. (1990) Regulation of the mevalonate pathway. Nature 343, 425–430 [DOI] [PubMed] [Google Scholar]
- 2. Brown M. S., and Goldstein J. L. (1980) Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J. Lipid Res. 21, 505–517 [PubMed] [Google Scholar]
- 3. Sharpe L. J., and Brown A. J. (2013) Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). J. Biol. Chem. 288, 18707–18715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DeBose-Boyd R. A. (2008) Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 18, 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goldstein J. L., DeBose-Boyd R. A., and Brown M. S. (2006) Protein sensors for membrane sterols. Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 6. Sever N., Yang T., Brown M. S., Goldstein J. L., and DeBose-Boyd R. A. (2003) Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell 11, 25–33 [DOI] [PubMed] [Google Scholar]
- 7. Liscum L., Finer-Moore J., Stroud R. M., Luskey K. L., Brown M. S., and Goldstein J. L. (1985) Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. J. Biol. Chem. 260, 522–530 [PubMed] [Google Scholar]
- 8. Roitelman J., Olender E. H., Bar-Nun S., Dunn W. A. Jr., and Simoni R. D. (1992) Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J. Cell Biol. 117, 959–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Song B. L., Sever N., and DeBose-Boyd R. A. (2005) Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell 19, 829–840 [DOI] [PubMed] [Google Scholar]
- 10. Jo Y., Lee P. C., Sguigna P. V., and DeBose-Boyd R. A. (2011) Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc. Natl. Acad. Sci. U.S.A. 108, 20503–20508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu T. F., Tang J. J., Li P. S., Shen Y., Li J. G., Miao H. H., Li B. L., and Song B. L. (2012) Ablation of gp78 in liver improves hyperlipidemia and insulin resistance by inhibiting SREBP to decrease lipid biosynthesis. Cell Metab. 16, 213–225 [DOI] [PubMed] [Google Scholar]
- 12. Hartman I. Z., Liu P., Zehmer J. K., Luby-Phelps K., Jo Y., Anderson R. G., and DeBose-Boyd R. A. (2010) Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J. Biol. Chem. 285, 19288–19298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morris L. L., Hartman I. Z., Jun D. J., Seemann J., and DeBose-Boyd R. A. (2014) Sequential actions of the AAA-ATPase valosin-containing protein (VCP)/p97 and the proteasome 19 S regulatory particle in sterol-accelerated, endoplasmic reticulum (ER)-associated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 289, 19053–19066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., and Brown M. S. (2002) Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110, 489–500 [DOI] [PubMed] [Google Scholar]
- 15. Goldstein J. L., and Brown M. S. (2015) A century of cholesterol and coronaries: from plaques to genes to statins. Cell 161, 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horton J. D., Goldstein J. L., and Brown M. S. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keller R. K., Zhao Z., Chambers C., and Ness G. C. (1996) Farnesol is not the nonsterol regulator mediating degradation of HMG-CoA reductase in rat liver. Arch. Biochem. Biophys. 328, 324–330 [DOI] [PubMed] [Google Scholar]
- 18. Edwards P. A., and Gould R. G. (1972) Turnover rate of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase as determined by use of cycloheximide. J. Biol. Chem. 247, 1520–1524 [PubMed] [Google Scholar]
- 19. Ness G. C., and Holland R. C. (2005) Degradation of HMG-CoA reductase in rat liver is cholesterol and ubiquitin independent. FEBS Lett. 579, 3126–3130 [DOI] [PubMed] [Google Scholar]
- 20. Engelking L. J., Liang G., Hammer R. E., Takaishi K., Kuriyama H., Evers B. M., Li W. P., Horton J. D., Goldstein J. L., and Brown M. S. (2005) Schoenheimer effect explained: feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J. Clin. Invest. 115, 2489–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., and DeBose-Boyd R. A. (2003) Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J. Biol. Chem. 278, 52479–52490 [DOI] [PubMed] [Google Scholar]
- 22. Simonet W. S., Bucay N., Lauer S. J., and Taylor J. M. (1993) A far-downstream hepatocyte-specific control region directs expression of the linked human apolipoprotein E and C-I genes in transgenic mice. J. Biol. Chem. 268, 8221–8229 [PubMed] [Google Scholar]
- 23. McFarlane M. R., Liang G., and Engelking L. J. (2014) Insig proteins mediate feedback inhibition of cholesterol synthesis in the intestine. J. Biol. Chem. 289, 2148–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jo Y., Sguigna P. V., and DeBose-Boyd R. A. (2011) Membrane-associated ubiquitin ligase complex containing gp78 mediates sterol-accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 286, 15022–15031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McDonald J. G., Smith D. D., Stiles A. R., and Russell D. W. (2012) A comprehensive method for extraction and quantitative analysis of sterols and secosteroids from human plasma. J. Lipid Res. 53, 1399–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mitsche M. A., McDonald J. G., Hobbs H. H., and Cohen J. C. (2015) Flux analysis of cholesterol biosynthesis in vivo reveals multiple tissue and cell-type specific pathways. Elife 4, e07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimano H., Horton J. D., Hammer R. E., Shimomura I., Brown M. S., and Goldstein J. L. (1996) Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J. Clin. Invest. 98, 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doolman R., Leichner G. S., Avner R., and Roitelman J. (2004) Ubiquitin is conjugated by membrane ubiquitin ligase to three sites, including the N terminus, in transmembrane region of mammalian 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for sterol-regulated enzyme degradation. J. Biol. Chem. 279, 38184–38193 [DOI] [PubMed] [Google Scholar]
- 29. Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., and Mangelsdorf D. J. (2000) Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 14, 2819–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Engelking L. J., Kuriyama H., Hammer R. E., Horton J. D., Brown M. S., Goldstein J. L., and Liang G. (2004) Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J. Clin. Invest. 113, 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DeBose-Boyd R. A., Ou J., Goldstein J. L., and Brown M. S. (2001) Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc. Natl. Acad. Sci. U.S.A. 98, 1477–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yabe D., Komuro R., Liang G., Goldstein J. L., and Brown M. S. (2003) Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 3155–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Engelking L. J., Evers B. M., Richardson J. A., Goldstein J. L., Brown M. S., and Liang G. (2006) Severe facial clefting in Insig-deficient mouse embryos caused by sterol accumulation and reversed by lovastatin. J. Clin. Invest. 116, 2356–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kita T., Brown M. S., and Goldstein J. L. (1980) Feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in livers of mice treated with mevinolin, a competitive inhibitor of the reductase. J. Clin. Invest. 66, 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reihnér E., Rudling M., Ståhlberg D., Berglund L., Ewerth S., Björkhem I., Einarsson K., and Angelin B. (1990) Influence of pravastatin, a specific inhibitor of HMG-CoA reductase, on hepatic metabolism of cholesterol. N. Engl. J. Med. 323, 224–228 [DOI] [PubMed] [Google Scholar]