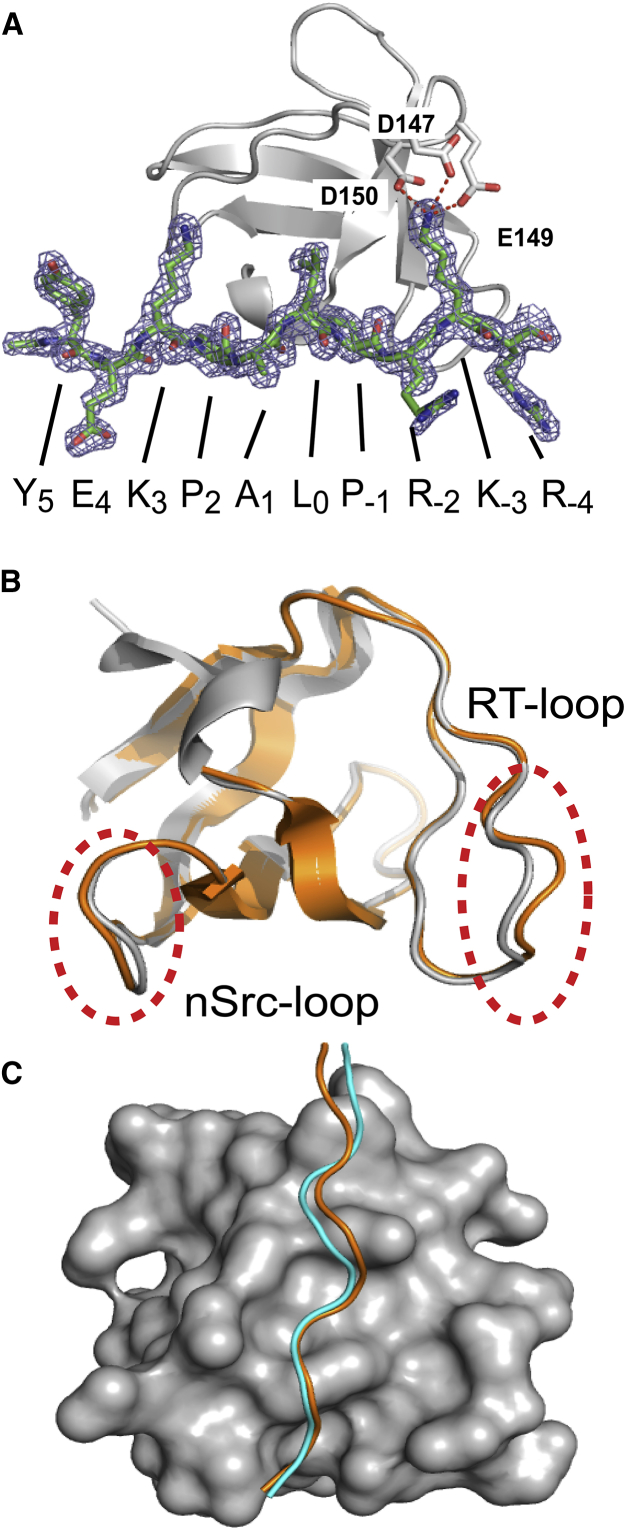
Figure 2.
The structure of the nSH3:PRM758 complex. (A) Representation of the 2Fo-Fc electron density map of PRM758 contoured at 1σ. The amino-acid sequence of PRM758 is shown at the bottom. Salt-bridges between K-3 of PRM758 and three acidic residues (D147, E149, and D150) of the nSH3 domain are shown in red. Salt-bridges between the acidic residues in nSH3 domain and K-3 of PRM758 are shown as dashed lines. Overlaid structures of (B) two nSH3 domains (gray and orange for molecules A and B in the PDB file, respectively) and (C) bound PRMs in an asymmetric unit of the nSH3:PRM758 complex crystal. PRMs bound to molecules A and B are shown in cyan and orange color, respectively. To see this figure in color, go online.