Summary
Quantitative high throughput cell array phenotyping (Q-HTCP) is applied to the genomic collection of yeast gene deletion mutants for systematic, comprehensive assessment of the contribution of genes and gene combinations to any phenotype of interest (phenomic analysis). Interacting gene networks influence every phenotype. Genetic buffering refers to how gene interaction networks stabilize or destabilize a phenotype. Like genomics, phenomics varies in its resolution with there being a tradeoff allocating a greater number of measurements per sample to enhance quantification of the phenotype vs. increasing the number of different samples by obtaining fewer measurement per sample. The Q-HTCP protocol we describe assesses 50,000–70,000 cultures per experiment by obtaining kinetic growth curves from time series imaging of agar cell arrays. This approach was developed for the yeast gene deletion strains, but it could be applied as well to other microbial mutant arrays grown on solid agar media. The methods we describe are for creation and maintenance of frozen stocks, liquid source array preparation, agar destination plate printing, image scanning, image analysis, curve fitting and evaluation of gene interaction.
Keywords: gene interaction, genetic buffering, quantitative high throughput cell array phenotyping (Q-HTCP), yeast mutant arrays, happy arrays, reference arrays, source arrays, dilution plates, destination plates, array map, perturbation, EASY Phenomics image analysis and growth curve fitting software
1. Introduction
Phenotypes are polygenic and mediated by gene interaction, meaning that genetic and environmental variables that influence a phenotype further depend on the allele status of multiple loci across the genome. This combinatorial aspect of the phenome renders it complex, and understanding this complexity is fundamental to mapping functional genomic variation (1). Saccharomyces cerevisiae is the best model for studying eukaryotic gene interaction due to the availability of yeast deletion mutant arrays, including deletions of non-essential genes and hypomorphic alleles of essential genes (2). To investigate genetic buffering networks, mutant strain cell arrays can be challenged with drugs or environmental variables and/or systematically combined with mutations of interest (3–8). Rigorous quantification of cell proliferation phenotypes enables accurate classification of gene interactions based on their strength of effect, thereby facilitating identification of genetic modules and buffering networks (3, 9, 10). Development of quantitative cell array phenotyping for growth curve analysis is in a relatively early stage (11), as its importance came to light only after work with the yeast mutant arrays, constructed fairly recently (12), revealed a high frequency of gene interaction (3, 6, 13, 14).
In a general sense, gene interaction is defined as the difference between the expected and observed phenotypes resulting from a genetic mutation in combination with another biological perturbation (see Section 3.6). Yeast mutant arrays provide genomic coverage of gene mutations, thus genetic buffering can be studied relatively comprehensively with respect to drug perturbation, environmental alteration, and/or introduction of additional mutations to study gene-drug, gene-environment, or gene-gene interaction, respectively. Q-HTCP is a protocol that involves cell array production, time-lapse imaging, image analysis, and fitting of the resulting data to a logistic growth function to obtain kinetic growth parameters that can be used to measure gene interaction across the entire genome (3, 15, 16). Quantification of gene interaction is also variable depending on a phenotypic parameter chosen for analysis, and a neutrality function that establishes an expected phenotype (17).
The methods described here are envisioned for collection of kinetic cell array phenotype data in a systematic and scalable way so that it can be analyzed in a flexible and cumulative manner. Gene interaction varies as a function of cellular context, and thus the interpretation of gene interaction networks depends on the biological nature of the aggregated phenotypic data (9). Q-HTCP can be used to obtain gene interaction profiles for any drug, environmental factor, or gene of interest. It is ideal if the perturbation of interest is rate limiting for cell proliferation so that both aggravating (enhancing) and alleviating (suppressing) interaction can be observed (16).
For gene-gene interaction studies, the synthetic genetic array (SGA) method, described elsewhere (6), should be carried out to construct double mutant strains before Q-HTCP. For application of the Q-HTCP method, the following variation of the SGA method may be considered: in the SGA method, double mutants are assessed for gene interaction in the final step of strain construction. SGA can be done quantitatively, where phenotypes are measured as colony outgrowth area at a selected time point, and a multiplicative neutrality function is used (6, 14, 18–20). However, if using Q-HTCP to quantify gene-gene interaction, a variation of the SGA procedure works better, where the mutation of interest is conditionally expressed, frozen stocks are made at the final step of SGA, and quantitative growth curve analysis is carried out as a subsequent step (16, 21). By utilizing conditional (e.g., tetracycline-regulated) expression of the query gene, interactions can be assessed at multiple levels of query gene expression by shifting cells to media containing different concentrations of tetracycline (22). Thus for Q-HTCP-based gene-gene interaction analysis, the SGA procedure is first used for strain construction (not phenotypic analysis) and subsequent experiments can incorporate a variety of media not compatible with SGA selection. Kinetic phenotyping and gradations of perturbation rigorously assess gene interaction, but in doing so, reduce the overall throughput in exchange for increased quantitative precision. Nevertheless, robust measurement of gene interaction can be performed on a phenomic scale (3, 15, 16, 21–23).
Future directions of Q-HTCP methodology include development of robotic cell array imaging equipment with supporting software (15, 18, 24, 25), database development for storing and retrieving Q-HTCP-derived gene interaction data (26), and network analysis for developing yeast phenomic models informative of human disease (16, 27).
2. Materials
Yeast mutant arrays consist of deletion strains for non-essential yeast genes (7, 12), hypomorphic alleles for essential genes called DAmP (Decreased Abundance by mRNA Perturbation) alleles where the 3′ untranslated region (UTR) of genes has been disrupted (5), and temperature sensitive alleles (4). In general, these are constructed in the same genetic background (e.g., BY4741, BY4742, or BY47473). These libraries provide nearly full coverage of the S. cerevisiae genome. The deletion strain project is described at: http://yeastdeletion.stanford.edu/. Strains can be purchased from Open Biosystems at Thermo Scientific. Q-HTCP methodology is applicable to any yeast mutant array, as well as other organisms or cell types that grow to a lawn on agar media.
Yeast media: Use YPD liquid media to passage cells and for making glycerol stocks (28), adding selective anti-biotics when useful (e.g., G418 and/or ClonNat/nourseothricin). It’s ideal to grow cell arrays (“happy arrays”) by inoculating from frozen glycerol stocks just prior to Q-HTCP. However, happy arrays can be stored at 4 degrees and used for a few weeks. The composition of the agar media is typically integral to the study design and thus experiment-specific. We use glass-distilled water, add amino acids after autoclaving, and use a Mediaclave (Integra Biosciences) and peristaltic pump for media consistency.
Pin tools: 96- and 384-pin tools can be obtained from V&P Scientific (http://www.vp-scientific.com/pin_tools.htm); ours were customized for use with the Sciclone ALH3000 instrument (Caliper Life Sciences). There are many types of pin tools, including manual versions and different robotic adapters. The main technical parameter is the diameter of the pin. For Q-HTCP, we use the FP6 style pin (1/16 inch cut with blunt end); these provide an adequate area to create a lawn when transferring cells from liquid to agar yet are small enough to accommodate a 384-format. A floating pin-style (not fixed/rigid) is best and is required for robotic printing.
Microtiter plates: Different microtiter plates are used throughout the protocol: a) “Rectangular monowell” plates (greater interior plate area) are used for imaging purposes (Nunc Cat# 267060) (see Note 1); b) “Notched monowell” plates (Nunc Cat# 242811) are preferred for SGA (because they are more affordable, provide orientation, and work well except for quantitative imaging), though the rectangular monowell plates also work; c) “Standard profile 384-well” plates (Nunc Cat# 242757) are used for glycerol stocks for long-term storage. These provide greater well volume- up to ~65 μL working volume- and are stackable; d) Low profile 384-well plates from Evergreen scientific (Evergreen Scientific Cat# 222-8210-01I and Cat# 290-8217-010) have cylindrical wells with a working volume of ~40 μL that yield more consistent volume of transfer, leading to better printing quality for the agar array. Therefore, Evergreen 384-well plates are used for “happy arrays” and “dilution arrays”, where precision in the volume of transfer affects the uniformity of the results; and e) “96-well plates” (Cat# 260860) are obtained from Nunc (see Note 2).
Tools for producing cell arrays: There are many robotics options, but manual array printing systems, with a mechanical guide to insure arrays are printed precisely the same, can also be used successfully. We primarily use the Sciclone ALH3000 (Automated Liquid Handling with 20-position work deck) integrated with Twister II (transport arm and microplate storage) (http://www.caliperls.com/products/lab-automation/). Such systems are operated by a PC, with control software (e.g., Caliper “Maestro” software), and provide a drag and drop graphical interface to create automated liquid handling methods.
Scanner for cell array imaging: The Epson Expression 10000 XL - Photo Scanner with Transparency Unit Lid.
Incubator: a floor model incubator is ideal given the high number (200 or more) of cell arrays one may wish to analyze simultaneously per experiment.
Centrifuge with swinging style rotor and microtiter plate adapters.
Plate Shaker: e.g., Lab-Line Instruments, Inc.; titer plate shaker model No. 4625.
3/32″ thick plexiglass cut to ½ ″ by 12″ and metal washers (for Scanner; see 3.4.3).
Adhesive aluminum foil (USA Scientific, Cat. # 2938-4100). We have found the foil rated for −40°C, rather than −80°C, prevents adhesive sticking to the plates when foil is removed.
‘Brayer’ from USA Scientific (http://www.usascientific.com/filmaccessories.aspx) for sealing glycerol stocks with adhesive foils.
Pin Cleaner: from V&P Scientific (http://www.vp-scientific.com/V&P_pincleaner.php)
For pin tool sterilization: 10% Bleach, sterile water, 100% ETOH, blotting paper, paint pad.
Roll of plastic wrap (e.g., Saran wrap).
Optional
-
15)
Bench top MediaClave - from Integra Biosciences (http://www.integra-biosciences.com/sites/mediaclave_new_1_e.html) facilitates media consistency through being programmable with continuous stirring and internal temperature monitoring.
-
16)
Peristaltic pump - streamlines pouring media from mediaclave. For use with the Mediaclave, a ‘homemade’ system for media pouring is sometimes helpful (available upon request).
-
17)
“Tough Tags” labels: these help at −80°C for reading writing on frosty plates; double labeling (on tag and directly on plate) is recommended for long-term storage.
3. Methods
3.1 Sterilizing the pin tool
The pin tool must be sterilized to avoid cross-contamination. Effective sterilization can be achieved by using 10% bleach to kill contaminating cells, water to rinse, and a final dip in ethanol followed by air drying (or ethanol with flaming if manually pinning). A ‘scrub pad’ (made from a paint pad obtained from a hardware store, cut to size of the microtiter plate; this can also be bought from V&P Scientific) is useful if working with cell paste transfers. The scrub pad should be wet with 10% bleach and is used at the start of the cleaning process to loosen the cell paste from the pins. Blotting paper is used to remove bleach before rinsing in water and to remove water before finally rinsing the pin tool in ethanol. Using two bleach baths provides additional sterilization, as cells will accumulate in the first bleach bath; 30–60 seconds at each step of the sterilization is adequate, and intervals can be empirically determined by experimental assessment of decontamination for a particular protocol.
3.2 Cleaning the pin tool
Whereas pin tool ‘sterilization’ is performed multiple times during every protocol, pin tool ‘cleaning’ is needed at the beginning of an entire experiment (e.g., every few days or so) to remove proteins, lipids and other debris that can build up on the pins. Prolonged use without cleaning can lead to loss of liquid transfer efficiency and uniformity. We use V&P Pintool cleaner (proprietary solution) and follow their directions. It’s a good habit to clean the pin tool before each Q-HTCP printing (we reuse the solution), and consider pintool cleaning anytime a decline or variability in spot quality is experienced. Sanding or polishing the pins may also improve the quality of cell array printing.
3.3 Creating and working with frozen glycerol cell array stocks
There are many scenarios for creating and handling cell array frozen stocks; examples are described below. When creating new cell arrays, (e.g., from hand-picking colonies off agar media, or to consolidate existing arrays through a “hit picking” process), it is generally best to first inoculate cultures in a 96-well liquid array format. By contrast, it is easier to replicate glycerol stocks already in 384 format by first growing the array on agar (e.g., similar to the final step of SGA). Slow growing cultures will reach a lower density on agar than in a liquid array, however very slow growing strains are difficult to work with regardless of how they are maintained. In the case of consolidating from 4 × 96-array format to 1 × 384-array format, propagating arrays in liquid or agar may work equally well. Whether on agar or in liquid, arrays are grown to near stationary phase (2–3 days) before glycerol stocks are made.
3.3.1. Create new 96-well-format cell array frozen stocks
Strains are provided commercially as 96-well glycerol stocks. To improve Q-HTCP throughput, strains should be consolidated into a 384-well format. Before consolidating however, making multiple copies of the library in 96-well format will help preserve it by providing a means of recovery in case of future cross contamination or other accidental loss. Multiple copies can be made at once; using a multi-channel pipette (robotic 96-channel if possible), draw an amount of volume of stock culture desired to inoculate all copies (e.g., 1–2 μL/copy) and distribute evenly. Alternatively, new 96-well arrays can be made by inoculating wells individually, such as consolidating hits for retesting and/or streaking the original library for single colonies to assess single-colony clones for reproducibility. After 2–3 days incubation at 30 degrees, autoclaved 80% glycerol is added to a final concentration of 20%; arrays are sealed with adhesive foil and shaken to mix glycerol and cells. To make copies:
Label and pre-fill plates with 150 μL (or less) YPD (G418 if appropriate for selection/sterility).
Re-suspend cultures of the existing 96-array (if copying an existing glycerol stock) and inoculate, or individually inoculate each well of the 96-well plate. Consider placing Reference Strains (ydl227c-Δ0::G418R) on each array when possible (see Notes 5 and 9).
Incubate at 30°C for 48–72 hrs.
Re-suspend cells by orbital shaking (this will facilitate final cell suspension after adding glycerol). Add sterile 80% glycerol to final concentration of 20% (50 μL of autoclaved 80% glycerol; use 80% glycerol to reduce viscosity).
Apply adhesive foil and affix with roller/brayer.
Mix by orbital shaking until the glycerol/water interface is gone and cells are evenly suspended. Pipetting is usually inadequate to suspend cells or mix glycerol. Store at −70°C immediately.
3.3.2 Consolidate 96-well frozen glycerol stock to 384-well arrays
To transfer yeast strain cultures from four 96-well plates into the corresponding quadrants of a 384-well plate, positions A1 from the first, second, third and fourth 96-well plates correspond (clockwise) to positions A1, A2, B2 and B1 on the 384-well plate, respectively. The working volume for 96 well plates is 100–200 μL, meaning cells can be suspended by orbital shaking without well spillover. Max volume for 96-well plates is 300 μL, thus after orbital shaking of cell arrays, additional volume can be added and suspension mixed by pipetting before transfer. The optimal volume for Evergreen plates is 30–40 μL or 40–65 μL for standard 384-well plate. Orbital shaking will not suspend cells or glycerol in 384 well plates, however this can be achieved by stirring with a pin tool. In general, glycerol should be mixed before adding to 384-well plate. 384-array glycerol stocks can be created simultaneously with making new 96 glycerol stocks (3.3.1), or by freezing/thawing 96 glycerol stocks. To consolidate from 96 to 384 arrays:
Prepare a sterile 20% solution of glycerol in water, YPD, or YPD supplemented with G418 if diluting/expanding existing stock prior to consolidation. Use 80% sterile glycerol if making new 96-well glycerol stocks and consolidating to 384-well at the same time (described in 3.3.1).
Thaw (if necessary) four 96-well frozen glycerol stock plates and shake to suspend cells, then remove foil (see Note 3).
Add to a 96-well plate a volume of 20% glycerol that will provide adequate volume to transfer to 384-well plates (e.g., 100 uL so that 50 uL can be transferred to each of two 384-well plates); more than one 384-well array can be made at a time. Mix by pipetting if well volume exceeds 200 μL (orbital shaking will cause spillover). Use a volume adequate to preserve the original 96-well stocks (slightly diluted) as the new 384-well stocks are created (see Note 4).
Seal plates with adhesive foil, and move promptly to −70°C to minimize time thawed. Retrieve the next set of 4 plates to thaw while next set of thawed plates are processed.
For downstream application, as a way of controlling for plate effects, add the reference strain (same genetic background with no gene deletion; we use the hoΔ0::G418R strain for this) to 3 of the 4 empty wells in Rows O and P of a 384 array. These are arranged in a square (e.g., O3, O4, P4, P3 for the MATa collection), resulting from an empty well on Row H of each 96-well array of deletion mutants. Leaving one of the four wells empty provides a way of distinguishing the MATa from a MATalpha library array (see Note 5).
3.3.3 Create/replicate frozen stocks from agar plates
This protocol is used to preserve double mutants after SGA, make additional copies of 384-array glycerol stocks, or as an alternative method of consolidating 96-well liquid arrays to a 384-well format (see Note 6).
Start by making a 384-agar cell array (e.g., endpoint of the SGA method or after inoculating liquid array to agar to replicate 384 stocks).
Label and pre-fill standard profile 384 well plate(s) with 60 μL YPD/20% glycerol.
Transfer cells with sterile pin tool directly from the agar media to the glycerol stock media (see Note 7).
Seal with adhesive aluminum foils, and store at −80°C.
Sterilize pin tool, and repeat steps above for each cell array stock.
3.4 Quantitative High Throughput Cell Array Phenotyping
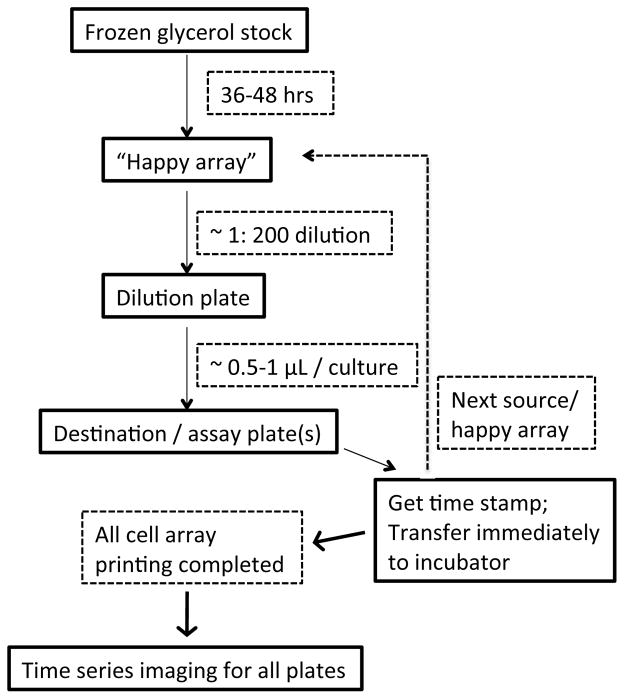
Overview: An ‘Experiment’, by definition, should have a completely matrixed design (Fig. 1); one axis of the matrix will be gene mutations (i.e., the identities of the strains on the yeast mutant arrays), and the other axis will be the perturbations systematically tested for every yeast mutant array. Perturbations can be drugs, genes (e.g. double mutants), or a combination (e.g., tetracycline-regulated gene in a query strain). Thus defined, multiple Q-HTCP experiments can be performed simultaneously, but the arrays should be grouped as separate experiments (do not place plates from different experiments on the same scan). Performing experiments in this way streamlines downstream analysis after the imaging step. Within an experiment, every source yeast mutant array will be tested with the same number and type of perturbations, which are defined by the make up of the media in the destination plates to which the yeast mutant arrays are transferred prior to imaging. Thus, the number of source arrays multiplied by the number of destination plate types is equal to the total number of arrays to be imaged as part of one experiment (see Notes 8 and 9).
Figure 1.
Q-HTCP workflow.
3.4.1 Making Happy Arrays
‘Happy arrays’ are freshly grown liquid culture arrays inoculated from glycerol stocks. Happy arrays increase reproducibility by reducing expression of confounding phenotypes, for example, gene-specific strain variability in freeze-thaw viability or survival in stationary phase. Happy arrays can be used after storage at four degrees, and uniform handling of the entire happy array collection can only help (e.g., time held at each temperature). It is not recommended to inoculate destination plates directly from glycerol stocks. To make happy arrays:
Label and fill Evergreen 384-well plates with 40 μL YPD (G418 or other selective antibiotic can be added to the media if desired to maintain mutant selection or prevent contamination). Consider filling a couple of extra plates for backup in case of error/need for extra plate during protocol.
Centrifuge filled plates 30 sec at ~400 × g (see Note 10).
Thaw cell array frozen stocks (groups of four are convenient to work with to minimize the time the arrays are out of freezer). Centrifuge arrays for 30 sec at ~400xg and remove foil (see Note 9).
Inoculate pre-filled Evergreen 384-well plates with glycerol stock (sterilize pin tool, stir pin tool on bottom of stock, transfer to happy array, repeat).
Reseal and return glycerol stocks to −80°C.
Wrap happy arrays with cellophane in packs of 5–10 plates to reduce evaporation and provide easier handling (Evergreen plates are not interlocking and do not stack well).
Incubate at 30°C for 48–72 hrs.
Optional: Observe happy arrays and record cultures that fail to grow. Alternatively, or in addition, vegetative growth can be assessed by printing to YPD agar as a control in the Q-HTCP experiment. Happy arrays can be stored at 4 degrees and used as short-term stocks (a few weeks).
3.4.2 Print happy arrays to test media
After they are fully-grown (48–72 hrs), happy arrays serve as ‘source’ arrays and are printed (i.e., transferred/inoculated) to ‘destination’ plates consisting of rectangular monowell plates filled with agar media. To optimally discriminate growth differences, a dilution step is recommended. The purpose of the ‘dilution’ plate is to reduce the cell density before transfer, similar to serial dilution in a traditional spot test. As diluted cultures go through more generations to reach the endpoint of growth, smaller differences in cell proliferation can be detected (3). From the dilution plate, the cell arrays are printed to agar media containing drugs or other variable of interest that constitute the assessment of gene interaction (see Note 11).
Prepare agar plates for an experiment and number them consecutively. The plates are printed in mini-series with each happy array being printed to all destination plate types (e.g. destination plates 1 thru 5= happy array source plate 1 printed to no-drug-control destination agar media, and media with high and low concentrations of Drug 1 and Drug 2; plates 6 thru 11 = happy array 2, plated to the next series of 5 destination plates, etc.). see Notes 8–12.
Prepare dilution plates by filling Evergreen 384-well plates with 40 μL sterile distilled water or media (it’s a good practice to make a couple of extra dilution plates). Media will usually yield larger spots than water (a given drop volume will spread out over a larger area); approximate volume to spot is .5–1 uL; approximate number of cells to spot is 200–1000 (see Notes 11 and 12).
Centrifuge dilution plates 30 sec at ~400g (to remove air bubbles; if forgotten, cultures may not be diluted correctly).
Transfer cells in series from the happy array to the dilution plate and then to the destination agar plates. As a rule of thumb, transfer ~0.5 μL from happy array to dilution plate (~1:100 dilution), and about ~.5 μL from destination to agar plate (~500 cells). Use pin tool to stir wells on source and dilution plates. Agar printing should be precise and reproducible with array squarely aligned with plate (use a robot or a mechanical guide, see Note 12).
Record the time that each happy array is processed in the workflow (this will be needed for growth curve fitting; the same time stamp can be used for all of the destination plates of a given source array as they are all moved to the incubator at the same time). Automated liquid handlers should have capability to collect print times, but this data must be recorded manually if not.
As the printing is completed for each set of agar destination plates corresponding to a happy array, move these to the incubator before processing the subsequent happy array (an important detail to equalize times of incubation for all arrays in the experiment as small temperature differences can have large effects on growth rate) (see Note 10).
3.4.3 Acquire images
-
1
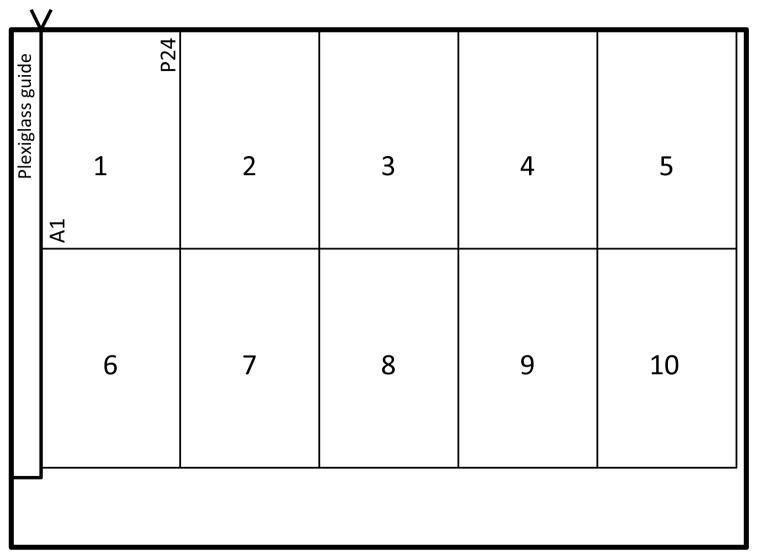
Use the Epson Expression 10000 XL - Photo Scanner with Transparency Unit Lid. Make sure a 3/32″ sheet of clear Plexiglas cut to ½ ″ × 12″ is used as a guide along the left side of the scanner, and that it is squarely flush against the glass border at top left. This is required to properly position the plates for imaging. The scanner transparency unit lid must be raised in the z-direction so that it can rest evenly across the plates; metal washers should be used for the purpose of holding the transparency unit lid in this optimal position. Remove the lids from the cell array agar plates prior to scanning. The scanner should accommodate 10 plates, oriented as shown in Fig. 2. Careful positioning of plates prior to scanning is essential, because the same array map will be used to analyze images from an entire experiment of scans (see Note 13).
Figure 2.
Q-HTCP scan layout.
Scanner settings
Mode: professional mode
Document type: Film
Film type: Positive film
Image type: 8-bit gray scale
Resolution: 280dpi
Document size: 12.20 in × 16.50 in
Target size: 12.20 in × 16.50 in
Scale: 100%
(uncheck all options):
No unsharp mask filter
No grain reduction
No dust removal
Scan File Type = Tiff
-
2
Create a new “experiment folder”, so that it can be selected from the scan driver as the location to save the images. The ‘film’ document type mode of scanning is used, in which the images if opened and viewed, will appear to be flipped horizontally, representing the view looking upward from beneath the glass surface of the scanner (this is normal, and the image analysis software assumes this orientation). Using the “positive film” mode, the cultures will appear gray and the background white. Begin scanning as soon as printing of all destination plates in an experiment is completed (this may be several hours after printing the first source to destination plate series). Scanning will be in groups of ten, and typically in the order the plates were printed. The file-naming scheme utilizes a format provided by the Epson scan driver and the resulting file names are important for proper calling by the image analysis software described below. In the Epson Scan driver, choose the “experiment folder” to store images. Name each scan set according to the approximate hours after the first scan set was taken. Use three digits followed by ‘hr’ (e.g., the first scan set will be ‘000hr’). The driver will automatically increment each scan file name and save to the selected experiment folder (e.g., 000hr001, 000hr002, etc.). The order for scanning and the order of plates on each scan must be maintained the same for every time point, and the placement of plates on the scanner must be precise for all scan sets throughout the experiment (Fig. 2). Scan sets should be acquired at 2–6 hour intervals for 3–5 days, until all cultures have reached their plateau phase of growth, or carrying capacity.
-
3
The first scan set (‘000hr’) should be obtained immediately after all plates are printed. Typically, the second scan set should be obtained 12–16 hrs after the first plates are printed if grown on glucose media (see Note 14). If spotting at higher density (e.g. without dilution), earlier scans will be needed; or alternatively, if the growth rate on the media (e.g., non-fermentable media) is slow, it may take longer before proliferation of the cell culture arrays is detectable by scanning. In any case, once the image threshold is reached (it generally correlates with when spots can be seen by careful visual inspection), scanning should be more frequent during early/exponential growth (e.g., once per doubling time), and can be less frequent as cultures begin reaching carrying capacity and growth slows down. It is ideal to have 6 or more readings during the dynamic growth of each culture. This will occur in different time windows on different scans, but the reference arrays serve as a guide (see Note 9).
Scanning protocol
Group plates in the incubator in sets of ten, in the order to be scanned. If performing more than one experiment at a time (see Note 8), start each new experiment as a separate scan set (i.e., do not combine plates from two different experiments on a single scan). Leave empty positions on the final scan of an experiment; do not fill them with plates from the subsequent experiment. Otherwise, always include ten cell arrays per scan.
Remove from the incubator one scan/stack of plates at a time (to avoid temperature effects).
Name scan images following the convention “000hr001” where the first 5 digits are entered into the Scan driver file name box and represent an approximate elapsed time from the first scan and the last three digits represent the scan number. The Epson scanner software will autoincrement and append the last digits as an auto-indexing feature (deselect the option to automatically overwrite file names).
Scans from all time points can go in the same experiment folder (e.g., “000hr001…. 000hr008; 010hr001… 010hr008, etc.). Each scan time series will be later separated prior to image analysis.
Continue scanning until all cultures appear to have reached a growth plateau at which time additional imaging is non-informative (see Note 15).
3.5 Kinetic analysis of growth from time series of cell array images
After imaging has been completed, image analysis and growth curve fitting is carried out using custom software. An earlier version of image analysis software for agar cell arrays was described (15). Time series images of agar cell arrays are analogous to OD readings of a liquid culture array. Compared to OD monitoring of liquid cultures, agar cell array image analysis reports over a greater dynamic range and does not saturate at high growth density; these data fit very well to a logistic growth function (15). For Q-HTCP the density of each spot culture is assessed as a pixel average over its area on the cell array. A newer version of the software (EASY Phenomics) takes multiple input files for image analysis (see Note 16). These include: 1) the raw images reorganized by placing each scan time series in a separate folder; 2) an array map file defining the approximate location of each spot culture on a scan (see Note 17); 3) a print times file, which defines ‘time zero’ when each proliferating agar cell array was created (by printing/inoculating from the happy array and dilution plate to the destination plate); and 4) the Master Plate and Drug/Media files, which contain information associated with each cell array in the experiment and their organization in the plate/scan layout. The latter information can be incorporated after image analysis and growth curve fitting has been satisfactorily completed; essentially providing the experimental labels for each growth curve. To perform Q-HTCP analysis with Easy Phenomics software (see Note 16):
-
Create the necessary files.
Create a directory of Q-HTCP experiments and an ‘Experiment Folder’ for each experiment. Sort the time series for each scan in the experiment into its own folder (000h001, 010hr001… etc. in a folder called “Scan1”; 000hr002, 010hr002… etc. in a folder called “Scan2”… etc.), and place all the Scan folders into the experiment folder.
Create a folder named ‘PT Map Scans’ within the Experiment Folder. Select a few good quality images, and copy them into the PT Map Scans folder. Use scan images containing cell arrays at all of the ten positions and where the plates are perfectly placed. Choose well-aligned scans without shifted or angulated plates, and with most spot cultures having growth (see Note 17).
-
Make a Print Times file, called ‘PrintTimes.txt’ and place it in the Experiment Folder. The print times file contains the time at which each Source Array was printed, marking the beginning of the growth curve for each set of destination plates; a single print time applies to all destination plates associated with a single source array. All plates associated with the same print time should be moved as a group to the incubator prior to printing the subsequent source array. The number of print times will be equal to the number of source arrays in the master plate file. The ‘PrintTimes.txt’ has the format:
3/1/2012 14.45
3/1/2013 14.52
-
Create a folder in the Experiment Folder called “MasterPlateFiles”. This folder contains the information about the Source Arrays and Destination Plates comprising the experiment, and will be incorporated to correctly label the cell array growth curve data (see Note 16).
Make a “Master Plate File”, consisting of the identities of strains on each source arrays file and store it in the ‘MasterPlateFiles’ folder (templates will be provided with the software).
Make a “DrugMedia” file and store it in the MasterPlateFiles folder (templates will be provided with the software). The DrugMedia file contains information about the media and perturbations that are being tested in the experiment (i.e., the series of destination plates that each source array is printed too).
-
After all files have been placed in their correct location in the Experiment Folder, run the EASY Phenomics software (see Note 16):
From the ‘File’ dropdown menu, select “New Analysis Folder” and browse to the correct Experiment Folder to create an Analysis Folder that will contain several folders used for image analysis, curve fitting, and report generation. The Analysis Folder will automatically be named with the current date with any additional name information input appended (the same experiment can be analyzed multiple times and ways). Results will be output to this Analysis Folder.
From the ‘Run’ drop down menu, select “Create PinTool Map”, and then select images from the ‘PT Map Scans’ folder to create an array map locating all 3840 spots on a scan. Inspect the array map and choose other scan images if needed to improve the array map, and continue when array map is satisfactory.
From the ‘Run’ drop down menu, select “Run Image Analysis”, and then choose one or more scan series to be analyzed. A pop up box will indicate when the image analysis is complete.
-
From the ‘Run’ drop down menu, select “Run Curve Fit”. The image density data for a single spot culture can be chosen and displayed as a plot with the fitted growth curve, or entire scans (3840 spot cultures per scan) can be analyzed in which case the growth parameters are output to a spreadsheet and saved in the Analysis Folder. For curve fitting, the data are fit to a logistic growth function:
G(t) is the size of the population at time, t. K is the carrying capacity, or final yield of the culture; r is the maximum specific rate (biologically, this is a population size-normalized rate, when there is nutrient excess); l is the time at which half the carrying capacity is reached, which also corresponds to the maximum growth rate (absolute population size increase, not normalized to population size). The parameters K, r, and l are cell proliferation phenotypes, which can be used to quantify interaction from multiple growth curves (15). A method for quantifying gene interaction is described in the next section, though the growth curve parameters could be employed in other models of gene interaction (16, 17).
-
Compile the final results.
From the “Generate Reports” drop down menu, select “Import MP and DrugMedia Data”, and browse to the MasterPlate and DrugMedia files in the MasterPlateFiles folder within the Experiment Folder and choose them. The data in these files will be associated with curve fits obtained previously.
From the “Generate Reports” drop down menus, select ‘Gen Results File’. This will incorporate all together, the labels, curve fit parameters, time series data used for curve fitting, statistical quality metrics and diagnostic criteria. The final result files are found in the PrintResults folder within the Experiment Analysis folder. This ResultsFile.txt should be saved as ResultsFile.xls file to further utilize the EASY Phenomics plotting and figure generation utilities (see Note 16).
3.6 Quantify Interactions
Conceptually, gene interactions provide experimental insight to predict how a phenotype resulting from biological perturbations can unexpectedly change when the perturbations occur in combination (1, 23). The biological perturbations must include at least one genetic mutation, where gene interaction can be generally represented as:
Phenotypeobserved is the directly-measured phenotype in the context of a perturbation combination (e.g., double mutant or single mutant additionally treated with drug). Phenotypeexpected incorporates the directly-measured phenotypes in the context of no perturbation and each single perturbation, and depends in part on a neutrality function (17), which reflects the assumptions inherent to determining the Phenotypeexpected (see Note 18). An approach we have recently found to be useful employs “ΔL” (l is a parameter of the logistic growth function known as Time to Maximum Growth (TMR), or the time at which half the carrying capacity (K/2) is achieved); it’s used as an indication of a time shift in the growth curve (15, 16). Using the l values to compare growth of cultures, we: a) determine the distribution of control strain phenotypes over a range of perturbation intensity; b) normalize the effect of the yeast mutant array strain mutation in the absence of a second perturbation (i.e., the single perturbations not treated with drug or dial down of a query gene); c) subtract the l value for each perturbation combination from the median single perturbation state of wild type strain; d) fit the differences between the single and double perturbation states across a range of perturbation intensity to a quadratic equation; and e) use the fitted difference at the highest perturbation intensity observed (16). Other neutrality models of interaction and/or incorporating other cell proliferation phenotypes (e.g., other parameters of the logistic function) can be investigated with Q-HTCP data. We expect that the utility of particular cell proliferation phenotypes (i.e., K, l, or r) will be different for different perturbations. For example, the phenotype induced by some drugs may be best reflected in the rate, while others are better reflected by the carrying capacity or L parameter.
With the goal of Q-HTCP providing growth curve-derived phenotypes that support high-resolution modeling of genetic buffering networks, it is important to establish protocols that can be applied across different laboratories will lead to an accumulation of gene interaction data that can be assessed in an integrative manner to advance our understanding of complex phenotypes in yeast, which may also be evolutionarily relevant for phenomic analysis of human disease (16).
3.7 Plate Sterilization
To help research funds go further and preserve the environment, plastic wear can be reused by scooping out agar and rinsing plates, soaking plates in 7% bleach in deionized water (with a dash of dishwashing liquid) for several hours, rinsing with deionized water, drying, rinsing with 100% ETOH, and storing in sealed plastic bins. (More details available upon request).
Acknowledgments
We thank funding agencies that have supported Q-HTCP-related work in the Hartman laboratory: Howard Hughes Medical Institute Physician-Scientist Early Career Award P/S ECA 57005927, American Cancer Society Research Scholar Grant RSG-10-066-01-TBE, Cystic Fibrosis Foundation Research Project MILLER08G0, NIH/NIA R01 AG043076. We thank collaborators who were supportive of Q-HTCP development including Eric Sorscher, Mary-Ann Bjornsti, Elizabeth Miller, Daniel Smith, Rick White, and Fritz Roth. We thank students who have worked to generate and analyze Q-HTCP, including Nic Tippery, Najaf Shah, Chandler Stisher, Crystal MaHarrey, Darryl Outlaw, Ray Louie, Robert Bone, Kyle Aune, Alexander Stepanov, and others.
Footnotes
Monowell plates should be labeled on the left side (A1 being in the top left corner), looking down on the agar array (see Figure 2), and the same volume of agar (38–40 mL) should be poured in every array. Use a striping pattern to indicate media type at time of pouring, but number plates consecutively just prior to cell array printing, when each source array will be printed to an identical series of different media types. Use 2% agar and allow it to cool ~45 minutes unlidded after pouring; this will improve printing by facilitating absorption of liquid after printing (see Note 8).
Except for the Evergreen Plates, we use all Nunc brand to maintain lid compatibility (lids are non-interchangeable between brands).
Work with 8–12 plates out of freezer at a time (in sets of four); one or two sets thawing while other is being transferred.
Rinse tips with water and sterilize by autoclaving to recycle/reuse and reduce cost.
In addition to placing the reference strain on each 384-well yeast mutant array to assess plate effects (e.g., mislabeling or other problem with media production), create full “Reference Arrays” (all wells have the reference strain) to assess position effects and print them at the beginning and end of each experiment. The Ref Arrays, if printed first and last in an experiment, provide assessment of technical variation from beginning to end of experiment, and also provide data to assess array position effects (e.g., variation in pin tool transfers or edge effects on cell proliferation). We recommend using the ho-Δ0 deletion strain (ydl227c-Δ0::G418R), which can also be manipulated identically to the yeast mutant arrays for SGA-construction of double mutant array controls (16).
Agar arrays are easier to work with, but slow growing strains may be outcompeted on the agar array (since the media is shared), thus yielding low-density stocks. Such strains can be difficult to analyze subsequently, however low density stocks can be partially restored to higher density at the “happy array” step of Q-HTCP (where media is not shared and each culture is grown to saturation). Since it is not possible to vortex in 384-well format, glycerol should be pre-mixed before adding to plates. Cells will not grow appreciably in the high glycerol media, so move glycerol stocks to −70°C storage soon after transferring cell paste.
Before freezing the new glycerol stock, consider inoculating fresh media for happy arrays that can be used to proceed directly to Q-HTCP without the need for a later freeze-thaw step.
Source-destination combinations should be unique within an “experiment” (no duplicates). For example if an experiment is being performed in replicate (e.g., every source to destination is printed in duplicate), replicate arrays should be grouped and scanned as two separate series, rather than a single ‘experiment’. In general, plates from each experiment are scanned together in successive groups of ten, and plates from different experiments should not be grouped on the same scan (if the last scan of one experiment contains less than ten plates, do not include the first plate of the subsequent experiment on that scan). For example, if in a single experiment there are 7 source arrays and 4 different destination/perturbation plates, with two replicates of each plate, the first replicate set would be imaged on scans 1–3, and the second replicate set on scans 4–6; scans 3 and 6 would consist of only 8 arrays.
Use a full “Reference Array” at the beginning and end of each experiment. The Ref Arrays provide assessment of technical variation from beginning to end of experiment and also provide data to assess ‘array position effects’. For the reference arrays, instead of the true reference strain (e.g., BY4741 or BY4742), which is functionally homothallic, we use the G418-sensitive ho-Δ0 deletion strain (ydl227c-Δ0::G418R), which can be manipulated identically to the yeast mutant arrays for growth on G418 and SGA-construction of double mutant array controls (16). In addition to the reference arrays, control strains can be added to 3 of the 8 empty positions on the yeast mutant arrays to aid identification of plate-to-plate variation among destination plates of the same type (see Note 5).
It is important to centrifuge the media-filled plates to remove trapped air bubbles that may occur during filling; if forgotten, bubbles can expand by trapping CO2 and forcing cultures out of well.
The exact amount of dilution matters less than consistency in the dilution factor for cultures across the array. Higher dilution enables discrimination between smaller growth differences, but it is important to have a lawn density high enough (100–1000 cells per spot) for confluent population growth, rather than distinct colonies, which introduce artifactual differences. Have the plates at room temp before printing and make sure the agar surface is dry. Printed cultures will run together where the agar surface is wet. To avoid this problem, allow agar plates to solidify with lid off (see Note 1), and store plates agar side up. It is ok to invert agar arrays after liquid drops have been absorbed into agar, but not before. Move sets of competed destination plates to incubator immediately and record print time before proceeding with cell array printing. If spots on destination arrays are not uniform, pin tool cleaning may help (3.2).
Using V&P Scientific FP6 (1/16″ diameter) style pins, 3 transfers will yield about the desired 0.5 μL. One can empirically determine the transfer volumes needed to obtain a light lawn of cells at each spot of the array. If there are spot quality/uniformity issues on the cell arrays, trouble-shooting includes cleaning the Pin tool with V&P cleaner or another suitable chemical wash. In addition, different agar media have different surface tension properties and water has greater surface tension than media. How well plates are dried also affects spot characteristics on array. If spots are not desirable size consider using media instead of water for dilution plates.
Create a directory to contain all Q-HTCP experiments. Within the Q-HTCP image folder, create a separate folder for each experiment and name accordingly (date, initials of experimenter, brief description of experiment, etc.). Scan 10 plates at a time unless it is the final scan of an experiment. Remove only 10 plates at a time from incubator. Pay careful attention to the placement of cell arrays on the scanner. Correctly position the plexiglass guide, and make sure all 10 cell arrays are flush against the glass surface border of the scanner, bases of plates are not overlapping, and all sides of arrays are squarely in contact (Fig. 2). Do not have plates from different experiments on the same scan. If working quickly, it will take ~2 minutes per scan; e.g., for experiment with 150 plates, expect it will take about 30 minutes to collect each time point of data.
Return plates quickly to incubator (only ten at a time should be out for scanning) to avoid plate effects due to temperature variation.
If plate contamination occurs during the course of scanning, in general it is easier to leave it alone, and remove the images prior to image analysis rather than trying to remove the contamination from the plate. If a plate must be removed during scanning, use an empty plate as a placeholder to maintain the scan positions for all other plates in the series. Keep a tray of water inside the incubator to increase humidity, which will slow cracking in the agar. Additionally, it may help to reduce airflow, depending on the incubator model.
The software is still under development, but we have compiled an executable prototype for use with this protocol. It is available upon request, along with a more detailed user’s manual.
Exemplary scan images from the experiment, where nearly all cultures on the scan are growing, should be used to create the array map. This is typically the reference arrays and arrays that are unperturbed (control media). Scans can be taken with ten exemplary cell arrays solely for the purpose of creating the array map file if there are not exemplary scans within the experiment. Multiple scans can be overlaid so that a composite image maps all cell array positions of a scan (see 3.5, step 2). The array map file may be similar from experiment to experiment, but it is good practice to obtain exemplary scan images specific for each experiment in case something about the cell array printing or placement of cell arrays changes from one experiment to another.
For quantifying interactions, it is recommended to incorporate summary statistics from the relevant cell proliferation parameter for the Reference Arrays to assess the full range of assay variation and to more confidently assess whether a particular gene mutant influences the phenotype.
Competing interests: JLH holds an equity interest in Spectrum PhenomX, LLC, which has a license to commercialize Q-HTCP technology. All other authors declare no competing interests.
References
- 1.Hartman JL, IV, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- 2.Dixon SJ, Costanzo M, Baryshnikova A, et al. Systematic mapping of genetic interaction networks. Annu Rev Genet. 2009;43:601–625. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- 3.Hartman JL, IV, Tippery NP. Systematic quantification of gene interactions by phenotypic array analysis. Genome Biol. 2004;5:R49. doi: 10.1186/gb-2004-5-7-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z, Vizeacoumar FJ, Bahr S, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011;29:361–367. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuldiner M, Collins SR, Thompson NJ, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 6.Tong AH, Evangelista M, Parsons AB, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 7.Winzeler EA, Shoemaker DD, Astromoff A, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 8.St Onge RP, Mani R, Oh J, et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo J, Tian D, McKinney BA, et al. Recursive expectation-maximization clustering: a method for identifying buffering mechanisms composed of phenomic modules. Chaos. 2010;20:026103. doi: 10.1063/1.3455188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartwell LH, Hopfield JJ, Leibler S, et al. From molecular to modular cell biology. Nature. 1999;402:C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 11.Blomberg A. Measuring growth rate in high-throughput growth phenotyping. Curr Opin Biotechnol. 2011;22:94–102. doi: 10.1016/j.copbio.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Giaever G, Chu AM, Ni L, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 13.Parsons AB, Brost RL, Ding H, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 14.Tong AH, Lesage G, Bader GD, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 15.Shah NA, Laws RJ, Wardman B, et al. Accurate, precise modeling of cell proliferation kinetics from time-lapse imaging and automated image analysis of agar yeast culture arrays. BMC Syst Biol. 2007;1:3. doi: 10.1186/1752-0509-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louie RJ, Guo J, Rodgers JW, et al. A yeast phenomic model for the gene interaction network modulating CFTR-ΔF508 protein biogenesis. Genome Med. 2012;4:103. doi: 10.1186/gm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mani R, St Onge RP, Hartman JL, IV, et al. Defining genetic interaction. Proc Natl Acad Sci U S A. 2008;105:3461–3466. doi: 10.1073/pnas.0712255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baryshnikova A, Costanzo M, Kim Y, et al. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods. 2010;7:1017–1024. doi: 10.1038/nmeth.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costanzo M, Baryshnikova A, Bellay J, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- 21.Singh I, Pass R, Togay SO, et al. Stringent Mating-Type-Regulated Auxotrophy Increases the Accuracy of Systematic Genetic Interaction Screens with Saccharomyces cerevisiae Mutant Arrays. Genetics. 2009;181:289–300. doi: 10.1534/genetics.108.092981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartman JL., IV Buffering of deoxyribonucleotide pool homeostasis by threonine metabolism. Proc Natl Acad Sci U S A. 2007;104:11700–11705. doi: 10.1073/pnas.0705212104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartman JL., IV . Genetic and Molecular Buffering of Phenotypes. In: Rodriguez R, Kaput J, editors. Nutritional Genomics: Discovering the Path to Personalized Nutrition. John Wiley & Sons; Hoboken, NJ: 2006. p. 496. [Google Scholar]
- 24.Collins SR, Schuldiner M, Krogan NJ, et al. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawless C, Wilkinson DJ, Young A, et al. Colonyzer: automated quantification of micro-organism growth characteristics on solid agar. BMC Bioinformatics. 2010;11:287. doi: 10.1186/1471-2105-11-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koh JL, Ding H, Costanzo M, et al. DRYGIN: a database of quantitative genetic interaction networks in yeast. Nucleic Acids Res. 2009;38:D502–507. doi: 10.1093/nar/gkp820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Califano A, Butte AJ, Friend S, et al. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet. 2012;44:841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burke D, Dawson D, Stearns T. Methods in Yeast Genetics. CSHL Press; 2000. [Google Scholar]