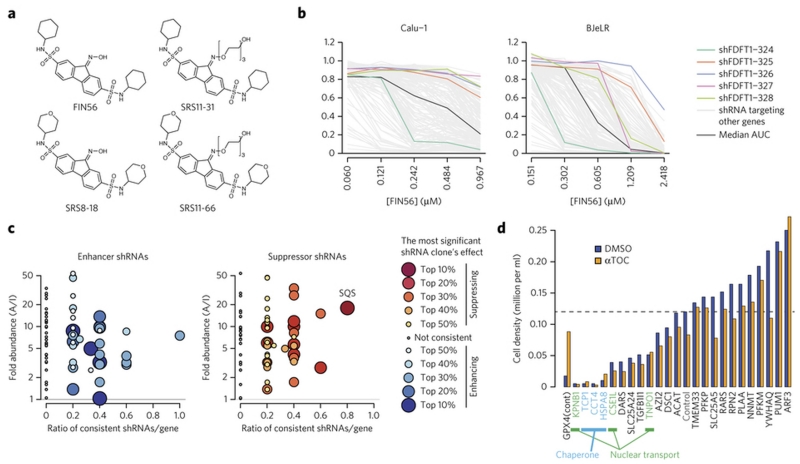
Figure 4. Squalene synthase (SQS) encoded by FDFT1 as FIN56’s target protein.
a. Active and inactive FIN56 analogs with PEG linkers. Supplementary Fig. 7 shows their potency in HT-1080 cells. b. Effects of five shRNAs against FDFT1 on FIN56. The results in two of the four cell lines were shown. Five shRNA clones targeting FDFT1 are shown in polychromatic lines. A black line indicates a shRNA which gives no effect (median AUC among tested shRNAs) in each cell line. Grey lines indicate shRNAs targeting other genes. c. Summary of proteomic target identification and shRNA screening targeting 70 identified genes on FIN56. Each dot summarizes the result of multiple shRNAs targeting a gene. Each shRNA is considered ‘consistent’ when it exerts indicated effect (enhancing or suppressing FIN56). X-axis shows the ratio: the number of consistent shRNAs inducing indicated (i.e., enhancing or suppressing FIN56) effects to the total number of shRNAs targeting the gene. Y-axis shows fold-enrichment of protein abundance on active versus inactive probe-beads in pull-down assay. See Supplementary Fig. 8 for more description. d. Effect of siRNAs against ‘loss-of-function’ candidates on BJeLR cell viability. Cells were grown under DMSO or ATOC (α-tocopherol)’s existence. shRNA screens in b,c were performed once in four cell lines. siRNA experiment in d was performed in BJeLR twice and mean of biological replicates.