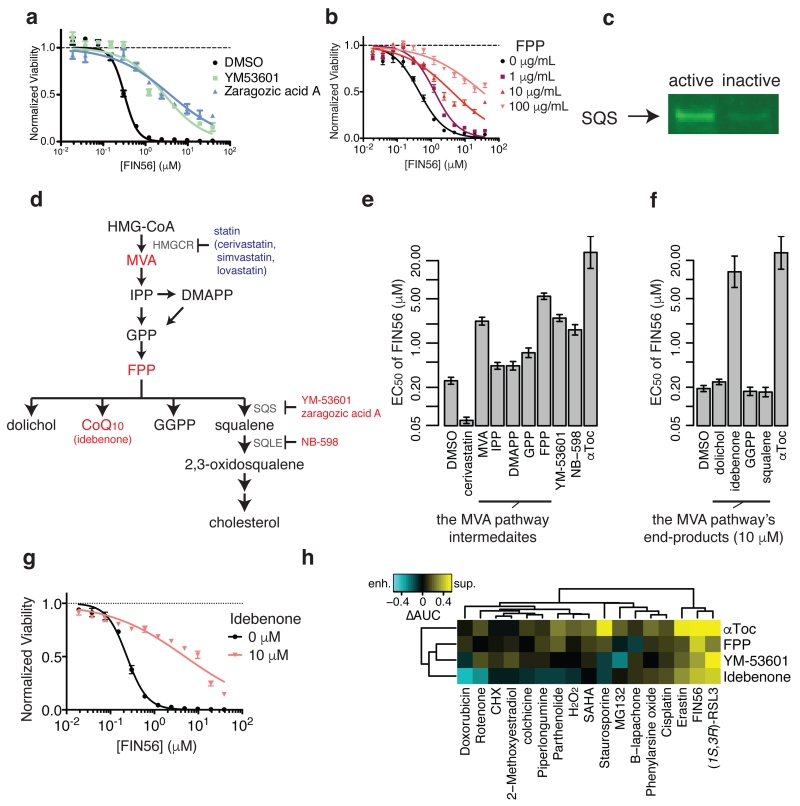
Figure 5. Validating SQS as the functionally relevant target to FIN56’s lethality.
a. Effects of chemical inhibitors of SQS on FIN56’s lethality. b. Effects of Farnesyl-PP on FIN56’s lethality. c. Detection of SQS using pull-down assay from HT-1080 whole cell lysate with active or inactive probes. Note that the probes are the same as what were used for chemoproteomic target identification. d. Schematics of the mevalonate pathway. Large letters are metabolites, small letters are enzymes responsible for the reactions or small molecules. Red and blue letters indicate the molecules (inhibitors or metabolites) suppressed or enhanced FIN56’s lethality. The detailed results are shown in e and f. e. Perturbation of the mevalonate pathway and their effects on FIN56’s lethality. Concentrations: cerivastatin (1 μM), metabolites (100 μM), YM-53601 (5 μM), NB-598 (25 μM), αToc (100 μM). f. Supplementation of 10 μM end-products of the MVA pathway and their effects on FIN56. g. Effect of 10 μM idebenone on FIN56 in HT-1080. h. Modulatory profiling between the modulators of the MVA pathway and various lethal compounds inducing oxidative stress. See Supplementary Fig. 9-–10 for SQS pull-down with competition and effect of statins on FIN56 lethality. a,b,g were performed in biological replicates and error-bars are s.e.m. of technical triplicates; e,f in biological duplicates and error-bars are standard errors of EC50 estimation from sigmoidal curve-fitting; h in singlicate. Full image of c is in Supplementary Figure 14.