Abstract
In 1882, Elie Metchnikoff identified myeloid-like cells from starfish larvae responding to the invasion by a foreign body (rose thorn). This marked the origins of the study of innate immunity, and an appreciation that cellular immunity is already well established in these “primitive” organisms. This chapter focuses on these myeloid cells as well as the newest members of this family, the dendritic cells (DC), and explores their evolutionary origins. Our goal is to provide evolutionary context for the development of the multilayered immune system of mammals, where myeloid cells now serve as central effectors of innate immunity and regulators of adaptive immunity. Overall, we find that core contributions of myeloid cells to the regulation of inflammation are based on mechanisms that have been honed over hundreds of millions of years of evolution. Using phagocytosis as a platform, we show how fairly simple beginnings have offered a robust foundation onto which additional control features have been integrated, resulting in central regulatory nodes that now manage multi-factorial aspects of homeostasis and immunity.
Keywords: myeloid cells, evolution, phagocytosis, immunity, inflammation, host-microbe interactions, homeostasis control, apoptosis, dendritic cell, follicular dendritic cell, lymphoid tissue
1. Introduction
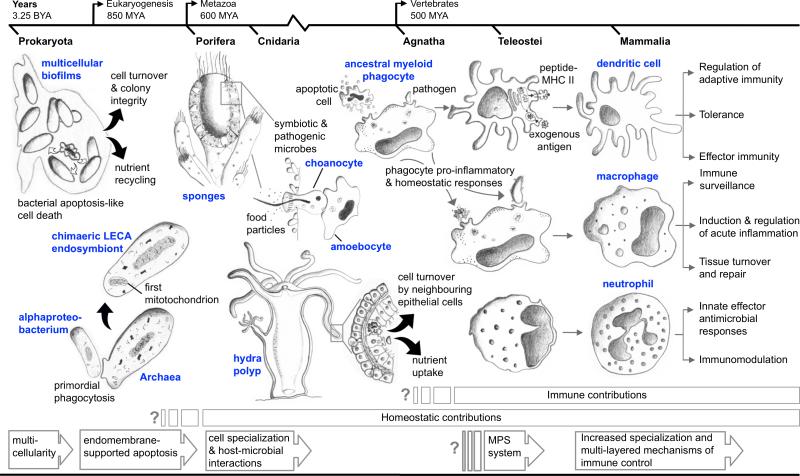
The drive to clear dying cells is evident even in the deepest branches of the Archea and Bacteria phylogeny, where colonial biofilms appeared as a mode to promote survival in diverse environments (1). These ancient colonies (earliest fossil record ~3.25 billion years ago) already displayed attributes of multi-cellular organism specialization, removing non-functional spent cells to recycle nutrients and maintain the integrity of the colony (1, 2). Multicellularity followed shortly after and introduced a requirement for removal of non-self (3, 4). Phagocytosis provided an elegant answer to both challenges, and has since served as a primary tool for cell turnover and removal of foreign invaders across all animal groups. It therefore provides a good stage to examine the evolution of myeloid cells through their contributions to homeostasis and host defenses (Figure 1). This chapter first focuses on ancestral phagocytes and examines their progression from primarily homeostatic cells to multifaceted effectors and regulators of immunity. The literature provides some insight into macrophage and lower metazoan hemocyte function as far back as echinoderms and urochordates. Further examination of gene marker conservation (e.g. apoptotic genes) in sponges and other colonial organisms allows us to dig deeper to examine the factors that led to the phylogenetic origins of cell clearance mechanisms and their continued evolution across newly developing animal branches. Subsequently, we focus on key challenges encountered by higher vertebrate myeloid cells as they manage increasingly more complex mechanisms of immunity while maintaining a strict balance between pro-inflammatory and homeostatic cellular responses. In one example, we examine the impact of specialization through the diverging contributions of macrophages and neutrophils. We then consider the continued specialization of the myeloid lineage through the eyes of the dendritic cell which, through antigen presentation, effectively integrated new adaptive features into well established and robust innate mechanisms of immunity. Indeed, documenting the multiple facets that comprise the life history of myeloid cells across evolution would not be possible in a single chapter. However, by focusing on their origins as phagocytes, we can appreciate the continued struggle of a host to develop novel and effective strategies to combat invading pathogens while ensuring the continued maintenance of tissue integrity and homeostasis.
Figure 1.
Contributions of phagocytosis to the evolution of myeloid cell function. Major events and key features identified in comparative models are highlighted. Metazoa refers to multicellular animals; invertebrates of the protostome lineage arose 600 million years ago (mya) and deuterostomes ~500 million years ago. Agnathans are jawless fish, and all other vertebrates have jaws (gnathostomes). Adaptive immunity is found only in the vertebrates, as well as the division of labor among myeloid cells that are well known in mammals. Refer to text for details of each of the particular features described in the figure.
2. Ancestral foundations for programmed cell death from prokaryotes
Biofilms represent large, complex bacterial communities that already share some of the challenges exhibited as eukaryotes manage multicellular structures. Ecologic opportunities within these colonies promote phenotypic diversity that enhances long-term population survival. As in eukaryotes, cell death allows for targeted turnover during development and removal of damaged cells following periods of stress (Figure 1) (2, 5). The bacterial programmed cell death process is coordinated through the action of holin-like proteins derived from cid and lrg operons, which were originally identified in Staphylococcus aureus but are widely conserved across Gram-positive and Gram-negative bacteria as well as Archea (2, 6). Functions for these proteins are analogous to the pro-apoptotic effector and anti-apoptotic regulators of the BCL-2 protein family (2). Their capacity to oligomerize in the bacterial membrane is also reminiscent of the oligomerization of effector BCL-2 proteins in the mitochondrial outer membrane, leading to membrane permeabilization and the release of cytochrome c during apoptosis (7). Additionally, similar to the manner by which eukaryotic pro-apoptotic BCL-2 effectors drive caspase activation and initiate the controlled demolition of cellular constituents, bacterial holin-like molecules activate peptidoglycan hydrolases that promote prokaryotic cell disassembly (6). Other bacterial effectors including RecA, ClpXP, BapE, the increased production of reactive oxygen species (ROS) and the SOS response further contribute to this process (8-10). These molecular events lead to downstream phenotypic features that are also consistent with a bacterial apoptosis-like cell death process based on observations of cell shrinkage, DNA fragmentation, chromosome condensation, extracellular exposure of phosphatidylserine and membrane depolarization (8, 9, 11, 12). Indeed, analyses of the evolution of apoptosis regulatory networks suggest that these already displayed a significant level of complexity prior to the development of the Metazoan line (13). As such, the seemingly altruistic behaviours from individual members of multicellular bacterial biofilms are simply a result of a cell turnover program that already shares several features of modern eukaryotic apoptotic mechanisms, ultimately contributing to the maintenance of colony integrity through management of cellular constituents after death.
Despite the parallels between eukaryotic apoptosis and apoptotic-like mechanisms in bacteria, however, there is little evidence that the latter is the preferred mode of death if biofilm homeostasis is to be maintained. Further, the lytic nature of this mechanism among bacteria is central to the release of genomic DNA that incorporates into the biofilm matrix to enhance biofilm integrity (14-16). The result is a colony with remarkable structural, mechanical and chemical properties that, among others, offers significant resistance to antimicrobial agents derived from other microorganisms in their environment or human intervention (17). This is markedly different from the classic inflammation paradigm of eukaryotes where cell lysis events contribute to pro-inflammatory rather than homeostatic tissue repair outcomes, based on the release of intracellular constituents (Figure 1). Thus, the mechanisms by which bacterial programmed cell death contributes to the integrity of the colony are quite distinct from those apoptotic-driven events that promote tissue repair and a return to homeostasis in multicellular eukaryotes.
3. The birth of the Eukaryota and endomembrane containment for apoptotic events
Phylogenetic analyses looking for conserved homologs encoding for phagocytosis-relevant proteins identified the early presence of these among a subset of archaea. Among others, these include actin nucleators that are monophyletic with eukaryotic players and further share unique structural features with the modern actin-related protein (ARP)-2/3 (18). The ARP2/3 complex is well established as a central regulator for the polymerization, organization and recycling of actin-filament networks (19). In phagocytosis, ARP2/3 is known to promote the formation of y-branched actin-filament networks that provide the structural integrity and mechanical force for lamellipodial protrusions and extension of the plasma membrane around the target particle. Results from phylogenomic analyses also point to the near universality of the actin-centered functional core across the Eukaryota, based on the conservation of nucleation-promoting factors WASP/N-WASP or WAVE/SCAR, as well as the actin-binding proteins gelsolin, profilin, cofilin, formin, and coronin that are involved in the remodeling of actin filaments (18). Their availability to the last eukaryotic common ancestor (LECA) would have offered the capacity to form branched-filament structures and networks during actin polymerization, an important feature for structural remodeling of the cell membrane that would then have facilitated bacterial uptake. This capacity for primordial phagocytosis appears to have been central to the establishment of endosymbionts where eubacterial and archaebacterial prokaryotic ancestors merged to establish a hybrid parent line for today's eukaryotes (Figure 1). The eubacterial target, presumably an alphaproteobacterium, subsequently became the first functional mitochondrion (20).
Despite continued debate over the morphological features for this archeal ancestor and the relative efficiency of its phagocytic machinery, support for this dominant model outlining the origins of Eukaryota can be found in cell biology, palaeontology and biochemical datasets (18, 20, 21). Additional features of contemporary eukaryotes that are proposed to have already been present in LECA are well reviewed elsewhere (20, 22). Overall, the chimaeric nature of this ancestral eukaryote offered new opportunities for further sophistication of the phagocytosis process beyond those which led to uptake of the bacterial endosymbiont. For example, phylogenomic analysis of the Ras superfamily of small GTPAses indicate that this group of important actin polymerization regulators was of bacterial origin - both eukaryotic and archaeal members are embedded within bacterial branches, and the cluster that include Rab-Ran-Ras-Rho GTPases was found to be overwhelmingly bacterial (18, 23). Thus, based on the symbiogenic model for the origin of the Eukaryota described above, horizontal gene transfer would have allowed for acquisition of novel genes from this family of GTPases from the phagocytosed eubacterial endosymbiont, further enriching the archeal parent genetic repertoire. As such, basal eukaryotes appear to have already possessed the capacity for phagocytosis and primordial mechanisms of programmed cell death that could help address early challenges associated with multicellularity (Figure 1).
Additional features acquired during eukaryogenesis further strengthened the platform for the long-standing evolutionary role that apoptosis and phagocytes play in homeostatic control. Among others, ancestral eukaryotes also gained containment of programmed cell death molecular events within membrane envelopes, which prevented the release of intracellular constituents (24). The emergence of this intricate endomembrane system is unique to eukaryotes (25). Based on this model, the eubacterial endosymbiont that evolved into the modern mitochondrion appears to have retained the capacity for autolysis as a mechanism to trigger downstream programmed cell death pathways. However, in light of the development of an endomembrane system, autolytic events now took place without the release of intracellular components that could be construed as danger signals to surrounding cells. Since acquisition of mitochondria appears to predate the origin of all known extant eukaryotes, current support points to a broad application for this novel mode of controlled cell death from the earliest days of the Eukaryota.
4. Host-microbial interactions and cellular specialization in basal multicellular metazoans
The sponges (phylum Porifera) are sessile, benthic aquatic organisms that represent the most basal clade of the extant multicellular animals (26). They lack specialized organs and a nervous system and thus must rely on the individual behavior of a limited number of cell types to manage their physiological requirements (27). For ancestral phagocytes, these contributions were based on meeting nutritional needs and managing emerging challenges with symbiotic and pathogenic microorganisms (Figure 1). Consistent with this idea, carnivorous sponges such as Asbestopluma hypogea already show the presence of specialized migrating cells that contribute to the digestion of disrupted macroprey (28). In other sponges, nutrients are derived from phagocytosis of free-living bacteria and other particles in the water column filtered through sponge choanocyte chambers (29, 30). As such, examples from the Porifera further support the early presence of the specialized cellular machinery necessary for chemotaxis, phagocytosis and intracellular degradation as well as their application in basal metazoans. Still, further research would help determine whether these archaeocytes (also called amoebocytes) and other sponge cells described to internalize food particles represent bonafide ancestral phagocytes that predate immune hemocytes and phagocytic myeloid cells of higher metazoans or an independent wave of cell specialization that permitted a shift in energy acquisition in the absence of a digestive cavity in deep-sea oligotrophic environments.
Notably, Porifera microbes comprise as much as 40% of the sponge volume, are well known to establish intimate relationships that range from mutualism to host-pathogen associations, and can be located in the host extracellular and intracellular spaces (31). This provides an opportunity to examine how early host-microbial associations may have led to the development of functional adaptations in basal metazoans and their interacting microorganisms. Not surprisingly, the most abundant bacterial morphotypes among microbial communities colonizing sponges display thickened cell walls, multiple membranes, and slime capsules, which appear to serve to counteract the phagocytic activity by sponge archaeocytes (32). Indeed, early analysis of discrimination among food bacteria (those utilized for nutrient acquisition) and bacterial symbionts showed selectivity in the recognition and uptake by sponge phagocytes (33). Whereas a large proportion of bacterial symbionts were seen to pass through the sponge and expelled in the exhalant current, food bacteria were retained within the sponge via phagocytic uptake. This is an extraordinary feat given the immense filtering capacity of sponges estimated at 24,000 L of water per day for a 1 kg sponge - such volume would translate to 2.4 × 1013 bacteria based on an estimated 106 bacteria per mL of sea water (34).
The selectivity of Porifera hosts for microbial targets described above is further complemented by well-developed bacterial strategies to survive intracellular killing mechanisms. Most recently, for example, symbionts from the sponge Cymbastela concentrica were found to contain a genomic fragment that encoded for four proteins, whose closest known relatives are part of the sponge genome (35). Two of these eukaryotic-like, ankyrin-repeat proteins (ARP) were found to interfere with phagosome development and acidification by blocking phagosome-lysosome fusion. Interestingly, ARPs are well-established tools for molecular mimicry of eukaryotic domains by bacterial pathogens like Legionella pneumophila and Coxiella burnetii, where they provide an effective mechanism to facilitate host infection and bacterial intracellular survival and proliferation (36-39). Thus, based on the characterization of modern sponges, both host and microbial contributions may have shaped the complexity of early phagocyte-microbial interactions in ancestral metazoans to determine if facultative or obligate symbiotic associations would promote colonization or whether effective rejection would result upon encounter of a pathogen.
5. Development of homeostatic pathways based on phagocytic uptake of apoptotic cells
Early work by Ellis and Horvitz in C. elegans identified genetic contributors with core regulatory roles in apoptotic events (40). C. elegans ced-4 was found to encode for a molecular chaperone necessary for the activation of ced-3 and promotion of apoptosis (41). In contrast ced-9 antagonized the actions of CED-3 and CED-4 in cells that should survive (42). We now know that these genes represent homologs for the mammalian apoptotic protease activating factor 1 (Apaf-1), interleukin-1-beta converting enzyme (ICE) and proto-oncogene bcl-2, respectively. More recent studies have extended the origin of this programmed cell death process into basal metazoans. Some of this work was described above. In addition, cnidarians like the Hydra have helped significantly to define the molecular events associated with apoptosis and the role of phagocytes in its homeostatic contributions in pre-bilaterians (Figure 1). This interplay has been observed during its regulation of cells numbers when feeding, tissue remodeling, gametogenesis (oogenesis and spermatogenesis), regeneration and allorecognition (24, 43, 44). In one example, apoptotic interstitial stem cells were rapidly internalized during normal growth and budding of hydra polyps by neighboring epidermal and gastrodermal epithelial cells. In another, hydra small nurse cells were internalized by surrounding oocytes during oogenesis. This process allowed for the efficient removal of spent cells and further supplied nourishment to developing polyps. The morphology of apoptotic Hydra cells is almost indistinguishable from that of higher metazoans (45). Further, the biochemical mechanisms mediating these events, the complexity of gene families contributing to them, as well as the intracellular localization and function of key players such as the caspase and Bcl-2 family proteins show a significant degree of conservation with those of higher metazoans (24, 43, 46). Thus, the contributions of phagocytes to the internalization and removal of apoptotic cells - defined as efferocytosis - is well established in cnidarians and appears to represent a platform mechanism for maintenance of homeostasis that arose in basal metazoans. Notably, the apoptosis cell death machinery of Hydra displays a significantly higher level of complexity than that of Caenorhabditis and Drosophila. This suggests that the primordial establishment of this novel homeostatic strategy was followed by selective gene loss after the divergence of the last common eumetazoan ancestor towards theprotostome clade rather than the previously suggested more dramatic evolution of the apoptotic machinery from worms to man based on the traditional Coelomata hypothesis that was in place at the time of early characterization of C. elegans apoptosis (47-50). This is consistent with examination of the apoptotic machinery in other “intermediate” metazoan clades such as the Mollusca, which also show higher level of complexity than Caenorhabditis and Drosophila (51). An additional point to highlight relates to the nature of phagocytes mediating the internalization of apoptotic cells in these basal metazoans. Despite the perceived dominance of mononuclear phagocytes for this uptake in mammals (tissue resident macrophages and immature dendritic cells) (52, 53), it appears that it was epithelial cells rather than bonafide professional phagocytes that were originally responsible for carrying out this process (Figure 1) (45). This may point to a segregation of labor towards the mononuclear phagocyte system that later evolved as these cells acquired a central position as inducers and regulators of inflammatory processes in higher metazoans. Alternatively, given that most studies on efferocytosis have focused largely on immune cell fractions and that apoptotic cells uptake and removal is highly efficient (1-2 hours), we may find that adherent epithelial populations actually play a dominant (rather than supportive) role in the execution of homeostatic cell clearance programs, and that this remains at least partially true in higher vertebrates. Off course, these studies will require further improvements in our current capacity to detect and quantitate the uptake of apoptotic cells in vivo in a range of cells and comparative animal models.
Apoptotic cell recognition by myeloid phagocytes initiates discrete molecular programs aimed at the promotion of homeostasis. In higher vertebrates, we find both the active induction of anti-inflammatory, wound-repair and tolerogenic mechanisms as well as the active repression of pro-inflammatory programs. Contributing factors include interleukin-10 (IL-10), transforming growth factor-β (TGF-β), platelet activating factor (PAF), prostaglandin E2, lipoxin A4, lactoferrin, vascular endothelial growth factor (VEGF) and others (54-56). For cytokines like IL-10 and TGF-β, and lipoxins like LXA4, this is mediated through a p38 MAPK-dependent mechanism and, at least in some cases, requires recognition of apoptotic cell phosphatidyl serine (57-59). Increased production of the factors above is accompanied by active translational control of pro-inflammatory mediators like interleukin-1 beta (IL-1β), interleukin- 6 (IL-6), interleukin-8 (IL-8) and tumor necrosis factor-alpha (TNF-α) (56). This is at least partially linked to repression of toll-like receptor (TLR) dependent and independent pro-inflammatory signaling cascades (56, 60). The production of ROS through the action of NADPH oxidase provides an additional regulatory node for the transition from pro-inflammatory to an anti-inflammatory homeostatic one (Figure 1). On the one hand, mammalian neutrophil ROS activates RhoA GTPase, a negative regulator of efferocytosis, in surrounding mature macrophages maintaining a pro-inflammatory phenotype and causing a reduction in apoptotic cell uptake (61). Conversely, inhibition of ROS production and RhoA activity increases efferocytosis efficiency, reduces TNF-α production, inhibits recruitment of inflammatory leukocytes, and promotes TGF-β release (62). This is consistent with experiments where the addition of exogenous apoptotic cells reduced neutrophil recruitment, promoted a shift in macrophages towards an anti-inflammatory phenotype, reduced ROS and pro-inflammatory cytokine production, and increased TGF-β expression (52, 63, 64). As such, phagocyte recognition of apoptotic cells offers key contributions for the modulation of inflammation from an innate immune mechanism that clearly discriminates between live and effete cells (65). Despite the complexity of these homeostatic programs, many of their components appear to have been already well established in prebilaterian metazoans. One good example is the TGF-β superfamily signaling pathway for which several components have already been identified, among others, in the ctenophore, Mnemiopsis leidyi, the sponge, Amphimedon, and the cnidarian, Trichoplax (66). However, we must be careful to not attribute inflammation regulatory roles to these pathways too early since, as with other components of the immune system, members of this network saw their origins not in immunity, but rather in the establishment of tolerance and tissue regeneration. Additional developmental functions described in both protostome and deuterostome models include tissue morphogenesis and dorsal-ventral patterning (67-69). Thus, it remains to be determined when exactly efferocytosis offered its first contributions to the control of pro-inflammatory immunity programs.
6. Core contributions of myeloid cells to the balance of pro-inflammatory and homeostatic responses in higher metazoans
Metchnikoff's seminal identification of phagocytosis in starfish larvae and an appreciation of its broader significance across various animal groups (Daphniae, snails, rhinoceros bettles, osseous fishes, frogs, turtles, lizards, pigeons, guinea pigs, rabbits, and others) provided strong early support for the prominent role of phagocytes in host defense and homeostasis across evolution. Unlike concurrent theories by Virchow and Cohnheim which focused on perenchymal and vascular responses, respectively, Metchnikoff saw leukocytes as the most active participants in an inflammatory response that was not a deleterious pathological process, but one that reflected a host's continued drive to maintain its integrity (70, 71):
It is possible to state as a general principle that the mesodermic phagocytes, which originally (as in the sponges of our days) acted as digestive cells, retained their role to absorb the dead or weakened parts of the organism as much as different foreign intruders (71).
More recent work has expanded Metchnikoff's early work in basal deuterostomes. It is now known, for example, that at least three distinct phagocyte morphotypes contribute to surveillance and immune defenses in larval and adult stages of echinoderms like the purple sea urchin (discoidal cells, polygonal cells and small phagocytes) (72). These, along with other echinoderm coelomocytes, further display nearly complete sets of homologs for vertebrate transcription regulators of myelopoiesis (e.g. PU.1/SpiB/SpiC/Ets) as well as key molecular players for pathogen recognition and uptake [e.g. greatly expanded gene families for Toll-like receptors (TLRs; >210 genes), Nod-like receptors (NLRs; >200 genes) and scavenger receptors (>1000 genes)]. Importantly, although “the attacked organism defended itself against these little aggressors by all the means at their disposal”, Metchnikoff also saw the internalization of neutrophils (aka microphage) by phagocytic macrophages as a key step in the subsequent resolution of inflammation (71). Indeed, it is now well established that phagocytes serve as ground zero for vertebrate induction and control of divergent pro-inflammatory and homeostatic responses following internalization of exogenous particles. At the site of infection, pathogen engagement leads to rapid production of pro-inflammatory mediators including reactive oxygen and nitrogen species (73, 74), antimicrobial peptides (75), and cytokines like TNF-α, IL-1β, and interferon gamma (IFN-γ) (76). The goal is to elicit potent responses that will deal with the incoming threat rapidly to prevent systemic dispersal of pathogens and additional commitment of energy resources. In contrast, internalization of apoptotic cells initiates a shift towards resolution mechanisms that promote tissue repair and a return to homeostasis once the pathogen has been effectively cleared. This is marked by increases in the production of transforming growth factor beta 1 (TGF-β1), interleukin 10 (IL-10), prostaglandin E2 and platelet activating factor (55, 77), combined with decreases in pro-inflammatory mediators, including TNF-α, leukotriene C4, thromboxane B2, and the interleukins IL-6, IL-8, IL-12, IL-17, and IL-23 (52, 78). Readers are further referred to the Inflammation and related chapters in this volume. Consistent with Metchnikoff's early ideas, this reflects a host's continued drive to maintain its integrity, and places myeloid phagocytes as central effectors and regulators of the inflammatory response. However, additional work will still be required to determine whether this dichotomy is a feature unique to vertebrates or also shared by invertebrate phagocytes.
Comparative examination of chordate myeloid populations shows the long-standing role that phagocytes have played in the induction and control of divergent pro-inflammatory and homeostatic responses. Like in mammals, professional phagocytes (macrophages, monocytes, neutrophils) are the primary contributors to the internalization of pro-inflammatory particles among teleost fish and agnathan hematopoietic leukocytes (79). Further, individual phagocytes of the jawless vertebrate Petromyzon marinus (sea lamprey), like those of teleost fish and mice, display the capacity for divergent pro-inflammatory and homeostatic responses following internalization of pathogens and apoptotic cells, respectively (63, 79). Despite these similarities, there are also preliminary indications of an evolutionary shift between the contributions of phagocytes to the induction versus the control of inflammation. When compared to their jawless counterparts, teleost phagocytes showed a greater capacity for internalization of pathogen-derived particles and for effective induction of pro-inflammatory antimicrobial programs. At the same time, higher vertebrate phagocytes displayed a lower sensitivity to homeostatic signals derived from apoptotic cell internalization (79). Thus, an early dominance of phagocyte homeostatic programs in evolution appears to have been followed by a progressive shift towards increased robustness among pro-inflammatory antimicrobial responses (Figure 1) (79). Of course, given the critical importance of inflammation control to the maintenance of tissue integrity and the management of precious metabolic resources it would seem ill-serving to favour the induction of pro-inflammatory host-defenses at the expense of inflammation control. Indeed, it appears that higher vertebrates have answered this dilemma through the establishment of new mechanisms of inflammation control among fist-line phagocytes, including the novel capacity of mammalian neutrophils to internalize apoptotic cells, a feature which is not available to their teleost fish counterparts (63, 80). Thus, an expanding body of evidence is showing that the contributions of mammalian neutrophils to inflammation control go well beyond their timely induction of apoptosis and subsequent removal by macrophages (81-83). An added division of labour now also sees important contributions to the control of inflammation beyond the myeloid professional phagocyte group (54, 82, 84, 85). However, as it is often the case, increased complexity is not always without problems; a quick scan of the literature shows extensive documentation of inflammation-driven disease among biomedical journals (86). An interesting possibility is that this may partially stem from a shorter evolutionary ‘break-in’ period for novel mechanisms of immune control that have only recently developed among the higher vertebrates.
8. Linking innate and adaptive immunity through dendritic cell specialization
With the appearance of adaptive immunity, the need for novel mechanisms of immune control in jawed vertebrates is obvious, not only with respect to the induction, regulation, and eventual resolution of inflammatory responses, but also with respect to the specific (and context-dependent) activation of antigen-restricted lymphocytes. The necessarily low precursor frequency of antigen-restricted lymphocytes generated by germline DNA rearrangement also necessitated specific anatomical locations (secondary lymphoid organs, SLO) in which both lymphocytes and antigen could be concentrated to facilitate the interaction of lymphocytes with cognate antigen. Additionally, a need arose for cells within the SLO capable of acquiring and presenting antigen to lymphocytes, as well as cells outside the SLO to acquire antigen and traffic it to the SLO for presentation.
The term “dendritic cell” was coined by Steinman and Cohn (87) to describe/define a morphologically distinct subset of cells in the SLO of mice,just over a century after the description of the archetypal dendritic cell (DC), the Langerhans cell, in human skin (Figure 1) (88, 89). In the years since their discovery, mammalian DC have been extensively characterized, and multiple subtypes with unique functions, tissue distributions, and surface phenotypes have been described (90). Macrophages and DC share multiple functional and phenotypic similarities (e.g. phagocytic capacity, MHC Class II expression) (Figure 1), as well as (in the case of conventional DC) a common progenitor/lineage (91). Despite the wealth of new and increasingly detailed information on DC, or perhaps because of it, the line between DC and macrophages has recently blurred (92). However, conventional DC can be functionally defined as cells with both a canonical dendritic morphology and, more importantly, the ability to capture and present antigen to antigen-restricted lymphocytes for the purpose of eliciting a specific adaptive immune response. DC with these properties have existed (at least) since the teleostei, and likely since the appearance of immunoglobulin (Ig)- and T cell receptor (TCR)/peptide:MHC (pMHC)-based adaptive immunity in the cartilaginous fish. Additionally, DC have been identified and studied with increasing detail in reptiles, amphibians, and birds. Herein, we hope to illustrate a progressive specialization in DC phenotype and function, alongside a retention of their ancestral and defining characteristics – MHC Class II expression and the ability to elicit specific adaptive immune responses – since their first appearance in early jawed vertebrates.
Adaptive immunity based on germline rearrangement of leucine-rich repeat gene segments, along with a T/B dichotomy, has recently been demonstrated in agnathans (93). However, as “there has been no sign of MHC I or II genes in animals older than the cartilaginous fish” (94), it seems unlikely that a canonical DC population (at least one partially defined by MHC II expression) exists in the jawless vertebrates. Adaptive immunity based on rearranging Ig-/TCR:pMHC arose in the jawed vertebrates, of which chondrichthyes is the oldest living group (cartilaginous fishes, i.e. chimeras, sharks, skates, and rays, common ancestor with humans ~500 MYA), as did the primordial secondary lymphoid organ, the spleen. Along with lymphocytes in the white pulp of the quiescent nurse shark (Ginglymostoma cirratum) spleen, we have described large cells expressing high levels of MHC Class II, and displaying dendritic processes (95). Immunization of adult nurse sharks with biotinylated bovine serum albumin resulted in an accumulation of immunogen in the splenic red pulp (RP) within one week; four weeks after immunization, the immunogen had localized to the splenic white pulp (WP), and was predominantly observed at the plasma membrane of large cells with dendritic processes (our unpublished observations). The only other putative DC population described in cartilaginous fish was identified in dogfish; Zapata and colleagues reported “irregular macrophages with long cell processes” in the hypothalamic ventricle (96), though their functional role in adaptive immunity was not addressed. Despite a lack of functional data for these putative DC in the nurse shark, their predicted role in the induction of adaptive immunity is supported by the appearance of not only MHC Class II but also CD83 (an immunoglobulin superfamily transmembrane protein associated with activated/differentiated DC) (97) in the nurse shark genome.
Some of the most definitive recent work on DC evolution has been performed in teleost fish (common ancestor with humans ~400 MYA), particularly in the zebrafish. Employing a cell sorting method based on a combination of a light scatter profile characteristic of myelomonocytes and binding of peanut agglutinin (PNA), a lectin which binds Gal-β(1-3)-GalNAc (a carbohydrate moiety present on, among other leukocytes, DC), Traver and colleagues (98) isolated cells from zebrafish whole kidney marrow, and demonstrated that these cells are bona fide DC by cellular morphology, expression of DC-associated transcipts (e.g. IL-12, MHC Class II invariant chain), and induction of antigen-restricted T cell proliferation. Further, they showed that these cells are present in peritoneal cavity, spleen, gut, thymus, and skin, though not in brain or liver (99), demonstrating that the zebrafish possesses not only cells with the phenotypic and functional characteristics of conventional DC, but also a tissue distribution pattern of these cells to some extent resembling that found in mammals.
In the Atlantic salmon (Salmo salar L.), a non-lymphocyte population of MHC Class II+, CD83+ (demonstrated by RT-PCR) cells with the ability to differentiate into cells with dendritic morphology has been identified (100). In the Rainbow trout (Oncorhynchus mykiss), cells with dendritic morphology have been identified as well, and were also demonstrated to be MHC Class II+, able to stimulate T cell proliferation in a mixed lymphocyte reaction, migratory, phagocytic, and had a gene expression profile of conventional DC (e.g. TLR, B7 family, CD83, CD209/DC-SIGN). In addition, like the cells identified in the salmon, the trout DC mature upon TLR simulation, increasing their expression of CD83 and MHCII, and elongating their dendritic processes (101). While these data do not formally identify a DC lineage distinct from that of macrophages, they do demonstrate a phenotypic and functional specialization strongly suggestive of canonical DC in teleost fish.
In the spleen of the amphibian Xenopus (common ancestor with humans ~350 MYA) “large, mitotically active cells with abundant electron lucent cytoplasm, large hyperlobated nuclei and prominent nucleoli are found in the periphery of the splenic white pulp” (102). These cells, termed XL cells, possess long cytoplasmic processes and, upon immunization, position themselves in a discrete ring at the internal perimeter of the WP, just inside the Grenzschichtenmembran of Sterba (a double layer of cells surrounding the WP and forming a boundary between it and the RP). XL cells have been implicated in the trafficking of antigen from the RP into the WP, and have been suggested to be capable of retaining native (i.e. unprocessed) antigen at their plasma membrane. Also upon immunization, native antigen is detectable at the internal perimeter of the WP, colocalizing with the immigrating/repositioning XL cells (103). Further, this antigen import into the WP was shown to be thymus-dependent – immunogen was undetectable in the WP of thymectomized animals. With these data in mind, XL cells were proposed to be a primitive follicular DC.
Follicular dendritic cells (FDC) were “originally identified by their striking morphology and their ability to trap immune complexes (ICs) of antigen and antibodies in B cell follicles” (104), and are instrumental in the initiation of humoral immune responses by virtue of their ability to retain and present native antigen, at their plasma membrane, to B cells. The precursor cells to FDC (pre-FDC) were recently identified as ubiquitous, perivascular mural cells expressing the platelet-derived growth factor receptor β (105), and as such these cells are of a non-hematopoietic lineage fundamentally distinct from both macrophages and conventional DC. Nonetheless, we feel that their inclusion in this review is necessary; FDC, or cells functionally comparable to FDC in their ability to trap antigen/ICs at their plasma membrane for presentation to B lymphocytes, are as central to the initiation of adaptive, humoral immune responses as conventional DC are to adaptive, cell-mediated immune responses. Additionally, the microarchitecture of the mammalian WP (characterized by a central arteriole surrounded by a periarteriolar sheath of T cells, with one or more B cell follicles adjacent, and all bounded by a marginal zone comprising a unique subset of B cells and two distinct subpopulations of macrophages) is dependent upon the lymphotoxin (LT) a1b2-dependent maturation of pre-FDC into mature FDC. Likewise, the establishment of germinal centers (GC) in mammals is both LT-dependent and FDC-dependent.
We have shown that the XL cells are indeed capable of acquiring and retaining native antigen at their plasma membrane after thymus-dependent immunization with phycoerythrin (PE) in Incomplete Freund's Adjuvant, suggesting the proposed FDC-like function. In addition, we have shown, both by flow cytometry and RT-PCR, that the XL cells express high levels of MHC Class II, which, along with the thymus-dependence of their antigen transport into the WP, suggests classical DC function as well (our unpublished observations). We have therefore proposed that the XL cells, the DC of the Xenopus spleen, perform “double duty,” presenting both native, surface-bound antigen to B cells as well as pMHC antigen to T cells. However, our data do not provide evidence for or against a distinct lineage of DC (separate from macrophages) in Xenopus. Nonetheless, the T cell-dependent migration of the Xenopus XL cells into the WP, as well as their positioning within the B cell follicle, demonstrate a functional maturation dependent upon (and a dedication to) interaction with lymphocytes suggestive of a specific role in the induction of adaptive immunity, distinct from their phagocytic capacity.
Outside the spleen (the only SLO in Xenopus), DC (as defined by morphology and MHC Class II expression) have been identified in both the thymus and skin (106, 107). Furthermore, cells in the skin with classical dendritic morphology and expressing vimentin have been identified, suggesting that the specialization of cells analogous to Langerhans cells occurred at the latest in the amphibians (108).
A population of cells similar to the Xenopus XL cells has been described in another amphibian, the Natterjack toad (Bufo calamata) (109). In this study, colloidal carbon particles injected into the dorsal lymph sac were first captured by macrophages and then transported into the RP, where cells with a similar morphology to Xenopus XL cells were observed. In a subsequent study (110), immunization with sheep red blood cells resulted in a similar appearance of antigen in the RP, followed by a progression of antigen into the WP, and also suggested that immunization resulted in the maturation/differentiation of monocytic cells into “giant, dendritic-like” cells (residing predominantly in the RP). As with the Xenopus XL cells, it is difficult to declare these cells distinct from macrophages; we hope that our current studies will help define the multiplicity vs. differentiation state of APC lineages in amphibians, and rekindle a general interest in the evolution of antigen presentation.
While reptiles (common ancestor with humans ~300 MYA) have been studied less extensively than amphibians, at least two examples of putative DC have been described. In the white pulp of the Asiatic reticulated python (Python reticulatus), a population of cells which, like the XL cells in Xenopus, localize to the periphery of the WP upon immunization, and are also able to trap and retain surface immunogen (111). In the Caspian turtle (Mauremys caspica), Zapata and colleagues originally described “dendritic macrophages” present within the reticular network of the WP (112), and later identified dendritic cells, characterized by dendritic morphology (with ultrastructural similarity to mammalian FDC) and weak phagocytic capacity, in the inner region of the periellipsoidal lymphoid sheath of the WP (113). Notably, the splenic WP of M. caspica comprises a central arteriole surrounded by a periarteriolar sheath of Ig-negative lymphocytes (presumably T cells), which is in turn surrounded by a periellipsoidal sheath, the inner layer of which contains Ig-positve B cells and the described dendritic cells. This is the earliest instance of splenic WP microarchitecture with remarkable similarity to the mammalian WP (though germinal centers have not been identified in any reptile).
Outside of mammals, DC subpopulations and lineages have been most extensively investigated in birds (common ancestor with humans ~200 MYA), particularly in the chicken Gallus gallus. Indeed, multiple subtypes of DC (in multiple anatomical locations) have been identified and characterized in the chicken (114). Langerhans cells in the esophagus and skin, identified by their cellular morphology, positioning within the epidermis, and expression of vimentin and MHC Class II, have been shown to be migratory from the epidermis to the dermal lymphoid nodules in response to hapten exposure. In the spleen, both ellipsoid-associated cells (similar to the cells described in the ellipsoid of the reptilian spleen) and their progeny, CD83-expressing interdigitating DC, are found in intimate contact with WP lymphocytes. In the bursa of Fabricius, bursal secretory dendritic cells (BSDC) are found. Each of these populations is CD45+, derived from hematopoietic precursors.
In addition to the DC subpopulations described above, Kaiser and colleagues have established a method of generating chicken bone marrow-derived DC (chBM-DCs) in vitro after culture in a combination of recombinant chicken GM-CSF and IL-4 (115). The cultured chBM-DC display the canonical dendritic morphology, are moderately phagocytic, capable of eliciting T cell proliferation in a mixed lymphocyte reaction, and express high levels of surface MHC Class II and CD11c, along with moderate levels of CD40. Upon stimulation with either LPS or CD40L, their phagocytic capacity decreases, surface expression of CD83 is acquired, and transcription of genes associated with induction of a canonical Th1 response is induced. Additionally, expression of the chemokine receptor CCR6 decreases concurrent with an increase in the expression of CCR7, indicating not only a functional maturation but also propensity toward migration/repositioning after activation (116).
Within the WP of the chicken spleen, cells with the canonical morphology, surface phenotype, and function of mammalian FDC have been identified. These chicken FDC are stellate, express VCAM-1 and ICAM-1 (among other cellular adhesion molecules), stain as positive for surface immunoglobulin (IgM and IgG/Y), and have a demonstrated capacity to stimulate both B cell proliferation and class switch recombination to IgG/Y without the ability to stimulate T cell proliferation (117). The lineage of these cells is controversial; conflicting reports regarding their expression of both CD45 and MHC Class II (positivity of each indicating a hematopoietic lineage) have been published. Regardless of the lineage of the chicken FDC, the microarchitecture of the chicken WP, like that of the reptile WP, is remarkably similar to that of mammals. In addition, germinal centers (GC, which in mammals are FDC-dependent) form in the chicken spleen. The microarchitectural organization of the chicken WP, along with the presence of GC, are surprising, given that the chicken genome has lost TNF-α, LT-α, and LT-β (118, 119), all of which are necessary for the establishment and maintenance of mammalian WP and GC.
The earlier years of the study of macrophages and dendritic cells naturally yielded differences between two cell types; a relative paucity of reagents for distinguishing the two made a distinction between them a natural starting point for experimentation. With the technological advances in multiparameter flow cytometry (and the dramatic increase in monoclonal antibodies to leukocyte markers) and transcriptomic analysis, it is almost inevitable that more and more similarities between cells with a common progenitor would be found. Nonetheless, we feel that DC are functionally distinct from macrophages in their propensity/dedication to the initiation of adaptive immune responses. With this almost purely functional distinction in mind, it is difficult to unequivocally identify the initial appearance of the DC – as a cell separate from the macrophage – in the evolution of adaptive immunity.
References
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol. 2007;5:721–6. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 3.Ereskovsky AV, Renard E, Borchiellini C. Cellular and molecular processes leading to embryo formation in sponges: evidences for high conservation of processes throughout animal evolution. Dev Genes Evol. 2013;223:5–22. doi: 10.1007/s00427-012-0399-3. [DOI] [PubMed] [Google Scholar]
- 4.Nedelcu AM. The evolution of self during the transition to multicellularity. Adv Exp Med Biol. 2012;738:14–30. doi: 10.1007/978-1-4614-1680-7_2. [DOI] [PubMed] [Google Scholar]
- 5.Lewis K. Programmed death in bacteria. Microbiol Mol Biol Rev. 2000;64:503–14. doi: 10.1128/mmbr.64.3.503-514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayles KW. Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol. 2014;12:63–9. doi: 10.1038/nrmicro3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ranjit DK, Endres JL, Bayles KW. Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J Bacteriol. 2011;193:2468–76. doi: 10.1128/JB.01545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bos J, Yakhnina AA, Gitai Z. BapE DNA endonuclease induces an apoptotic-like response to DNA damage in Caulobacter. Proc Natl Acad Sci U S A. 2012;109:18096–101. doi: 10.1073/pnas.1213332109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dwyer DJ, Camacho DM, Kohanski MA, Callura JM, Collins JJ. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol Cell. 2012;46:561–72. doi: 10.1016/j.molcel.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wadhawan S, Gautam S, Sharma A. Metabolic stress-induced programmed cell death in Xanthomonas. FEMS Microbiol Lett. 2010;312:176–83. doi: 10.1111/j.1574-6968.2010.02114.x. [DOI] [PubMed] [Google Scholar]
- 11.Bidle KD, Falkowski PG. Cell death in planktonic, photosynthetic microorganisms. Nat Rev Microbiol. 2004;2:643–55. doi: 10.1038/nrmicro956. [DOI] [PubMed] [Google Scholar]
- 12.Hakansson AP, Roche-Hakansson H, Mossberg AK, Svanborg C. Apoptosis-like death in bacteria induced by HAMLET, a human milk lipid-protein complex. PLoS One. 2011;6:e17717. doi: 10.1371/journal.pone.0017717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zmasek CM, Godzik A. Evolution of the animal apoptosis network. Cold Spring Harb Perspect Biol. 2013;5:a008649. doi: 10.1101/cshperspect.a008649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conover MS, Mishra M, Deora R. Extracellular DNA is essential for maintaining Bordetella biofilm integrity on abiotic surfaces and in the upper respiratory tract of mice. PLoS One. 2011;6:e16861. doi: 10.1371/journal.pone.0016861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 17.Schultz D, Onuchic JN, Ben-Jacob E. Turning death into creative force during biofilm engineering. Proc Natl Acad Sci U S A. 2012;109:18633–4. doi: 10.1073/pnas.1215227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yutin N, Wolf MY, Wolf YI, Koonin EV. The origins of phagocytosis and eukaryogenesis. Biol Direct. 2009;4:9. doi: 10.1186/1745-6150-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–26. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 20.McInerney JO, O'Connell MJ, Pisani D. The hybrid nature of the Eukaryota and a consilient view of life on Earth. Nat Rev Microbiol. 2014;12:449–55. doi: 10.1038/nrmicro3271. [DOI] [PubMed] [Google Scholar]
- 21.Martijn J, Ettema TJ. From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochem Soc Trans. 2013;41:451–7. doi: 10.1042/BST20120292. [DOI] [PubMed] [Google Scholar]
- 22.Poole AM, Neumann N. Reconciling an archaeal origin of eukaryotes with engulfment: a biologically plausible update of the Eocyte hypothesis. Res Microbiol. 2011;162:71–6. doi: 10.1016/j.resmic.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Elias M, Klimes V. Rho GTPases: deciphering the evolutionary history of a complex protein family. Methods Mol Biol. 2012;827:13–34. doi: 10.1007/978-1-61779-442-1_2. [DOI] [PubMed] [Google Scholar]
- 24.David CN, Schmidt N, Schade M, Pauly B, Alexandrova O, Bottger A. Hydra and the evolution of apoptosis. Integr Comp Biol. 2005;45:631–8. doi: 10.1093/icb/45.4.631. [DOI] [PubMed] [Google Scholar]
- 25.Jekely G. Small GTPases and the evolution of the eukaryotic cell. Bioessays. 2003;25:1129–38. doi: 10.1002/bies.10353. [DOI] [PubMed] [Google Scholar]
- 26.Thacker RW, Diaz MC, Kerner A, Vignes-Lebbe R, Segerdell E, Haendel MA, Mungall CJ. The Porifera Ontology (PORO): enhancing sponge systematics with an anatomy ontology. J Biomed Semantics. 2014;5:39. doi: 10.1186/2041-1480-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergquist PR. Sponges. University of California Press; Berkeley, CA.: 1978. [Google Scholar]
- 28.Vacelet J, Duport E. Prey capture and digestion in the carnivorous sponge Asbestopluma hypogea (Porifera : Demospongiae). Zoomorphology. 2004;123:179–190. [Google Scholar]
- 29.Wehrl M, Steinert M, Hentschel U. Bacterial uptake by the marine sponge Aplysina aerophoba. Microbial Ecology. 2007;53:355–365. doi: 10.1007/s00248-006-9090-4. [DOI] [PubMed] [Google Scholar]
- 30.Hadas E, Shpigel M, Ilan M. Particulate organic matter as a food source for a coral reef sponge. J Exp Biol. 2009;212:3643–3650. doi: 10.1242/jeb.027953. [DOI] [PubMed] [Google Scholar]
- 31.Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol Mol Biol R. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, Hacker J, Moore BS. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol. 2002;68:4431–40. doi: 10.1128/AEM.68.9.4431-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkinson CR, Garrone R, Vacelet J. Marine sponges discriminate between food bacteria and bacterial symbionts - electron-microscope autoradiography and insitu evidence. Proc R Soc B. 1984;220:519–528. [Google Scholar]
- 34.Vogel S. Current-induced flow through living sponges in nature. Proc Natl Acad Sci U S A. 1977;74:2069–71. doi: 10.1073/pnas.74.5.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen MT, Liu M, Thomas T. Ankyrin-repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol Ecol. 2014;23:1635–45. doi: 10.1111/mec.12384. [DOI] [PubMed] [Google Scholar]
- 36.Habyarimana F, Al-Khodor S, Kalia A, Graham JE, Price CT, Garcia MT, Kwaik YA. Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ Microbiol. 2008;10:1460–74. doi: 10.1111/j.1462-2920.2007.01560.x. [DOI] [PubMed] [Google Scholar]
- 37.Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320:1651–4. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price CT, Al-Khodor S, Al-Quadan T, Abu Kwaik Y. Indispensable role for the eukaryotic-like ankyrin domains of the ankyrin B effector of Legionella pneumophila within macrophages and amoebae. Infect Immun. 2010;78:2079–88. doi: 10.1128/IAI.01450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Schaik EJ, Chen C, Mertens K, Weber MM, Samuel JE. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat Rev Microbiol. 2013;11:561–73. doi: 10.1038/nrmicro3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–29. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 41.Yuan J, Horvitz HR. The Caenorhabditis elegans cell death gene ced-4 encodes a novel protein and is expressed during the period of extensive programmed cell death. Development. 1992;116:309–20. doi: 10.1242/dev.116.2.309. [DOI] [PubMed] [Google Scholar]
- 42.Hengartner MO, Ellis RE, Horvitz HR. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature. 1992;356:494–9. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- 43.Reiter S, Crescenzi M, Galliot B, Buzgariu W. Hydra, a versatile model to study the homeostatic and developmental functions of cell death. Int J Dev Biol. 2012;56:593–604. doi: 10.1387/ijdb.123499sr. [DOI] [PubMed] [Google Scholar]
- 44.Burnet FM. “Self-recognition” in colonial marine forms and flowering plants in relation to the evolution of immunity. Nature. 1971;232:230–5. doi: 10.1038/232230a0. [DOI] [PubMed] [Google Scholar]
- 45.Bottger A, Alexandrova O. Programmed cell death in Hydra. Semin Cancer Biol. 2007;17:134–46. doi: 10.1016/j.semcancer.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Lasi M, David CN, Bottger A. Apoptosis in pre-Bilaterians: Hydra as a model. Apoptosis. 2010;15:269–78. doi: 10.1007/s10495-009-0442-7. [DOI] [PubMed] [Google Scholar]
- 47.Edgecombe GD, Giribet G, Dunn CW, Hejnol A, Kristensen RM, Neves RC, Rouse GW, Worsaae K, Sorensen MV. Higher-level metazoan relationships: recent progress and remaining questions. Org Divers Evol. 2011;11:151–172. [Google Scholar]
- 48.Holland PWH. The future of evolutionary developmental biology. Nature. 1999;402:C41–C44. doi: 10.1038/35011536. [DOI] [PubMed] [Google Scholar]
- 49.Aravind L, Dixit VM, Koonin EV. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291:1279–84. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- 50.Vaux DL, Strasser A. The molecular biology of apoptosis. Proc Natl Acad Sci U S A. 1996;93:2239–44. doi: 10.1073/pnas.93.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sokolova IM. Apoptosis in molluscan immune defense. Invertebrate Surviv J. 2009;6:49–58. [Google Scholar]
- 52.Poon IKH, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nature Rev Immunol. 2014;14:166–180. doi: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henson PM, Hume DA. Apoptotic cell removal in development and tissue homeostasis. Trends Immunol. 2006;27:244–50. doi: 10.1016/j.it.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Ortega-Gomez A, Perretti M, Soehnlein O. Resolution of inflammation: an integrated view. EMBO Mol Med. 2013;5:661–74. doi: 10.1002/emmm.201202382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fadok VA, Chimini G. The phagocytosis of apoptotic cells. Sem Immunol. 2001;13:365–372. doi: 10.1006/smim.2001.0333. [DOI] [PubMed] [Google Scholar]
- 56.Elliott MR, Ravichandran KS. Clearance of apoptotic cells: implications in health and disease. J Cell Biol. 2010;189:1059–1070. doi: 10.1083/jcb.201004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung EY, Liu J, Homma Y, Zhang Y, Brendolan A, Saggese M, Han J, Silverstein R, Selleri L, Ma X. Interleukin-10 expression in macrophages during phagocytosis of apoptotic cells is mediated by homeodomain proteins Pbx1 and Prep-1. Immunity. 2007;27:952–64. doi: 10.1016/j.immuni.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hottz ED, Medeiros-de-Moraes IM, Vieira-de-Abreu A, de Assis EF, Vals-de-Souza R, Castro-Faria-Neto HC, Weyrich AS, Zimmerman GA, Bozza FA, Bozza PT. Platelet activation and apoptosis modulate monocyte inflammatory responses in dengue. J Immunol. 2014;193:1864–1872. doi: 10.4049/jimmunol.1400091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kronke G, Katzenbeisser J, Uderhardt S, Zaiss MM, Scholtysek C, Schabbauer G, Zarbock A, Koenders MI, Axmann R, Zwerina J, Baenckler HW, van den Berg W, Voll RE, Kuhn H, Joosten LA, Schett G. 12/15-lipoxygenase counteracts inflammation and tissue damage in arthritis. J Immunol. 2009;183:3383–9. doi: 10.4049/jimmunol.0900327. [DOI] [PubMed] [Google Scholar]
- 60.Pattabiraman G, Lidstone EA, Palasiewicz K, Cunningham BT, Ucker DS. Recognition of apoptotic cells by viable cells is specific, ubiquitous, and species independent: analysis using photonic crystal biosensors. Mol Biol Cell. 2014;25:1704–1714. doi: 10.1091/mbc.E13-11-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McPhillips K, Janssen WJ, Ghosh M, Byrne A, Gardai S, Remigio L, Bratton DL, Kang JL, Henson P. TNF-alpha inhibits macrophage clearance of apoptotic cells via cytosolic phospholipase A2 and oxidant-dependent mechanisms. J Immunol. 2007;178:8117–26. doi: 10.4049/jimmunol.178.12.8117. [DOI] [PubMed] [Google Scholar]
- 62.Moon C, Lee YJ, Park HJ, Chong YH, Kang JL. N-acetylcysteine inhibits RhoA and promotes apoptotic cell clearance during intense lung inflammation. Am J Respir Crit Care Med. 2010;181:374–87. doi: 10.1164/rccm.200907-1061OC. [DOI] [PubMed] [Google Scholar]
- 63.Rieger AM, Konowalchuk JD, Grayfer L, Katzenback BA, Havixbeck JJ, Kiemele MD, Belosevic M, Barreda DR. Fish and mammalian phagocytes differentially regulate pro-inflammatory and homeostatic responses in vivo. PLoS One. 2012;7:e47070. doi: 10.1371/journal.pone.0047070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rieger AM, Havixbeck JJ, Belosevic M, Barreda DR. Teleost soluble CSF-1R modulates cytokine profiles at an inflammatory site, and inhibits neutrophil chemotaxis, phagocytosis, and bacterial killing. Dev Comp Immunol. 2015;49:259–66. doi: 10.1016/j.dci.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 65.Birge RB, Ucker DS. Innate apoptotic immunity: the calming touch of death. Cell Death Differ. 2008;15:1096–102. doi: 10.1038/cdd.2008.58. [DOI] [PubMed] [Google Scholar]
- 66.Pang K, Ryan JF, Baxevanis AD, Martindale MQ. Evolution of the TGF-beta signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi. PLoS One. 2011;6:e24152. doi: 10.1371/journal.pone.0024152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Robertis EM, Sasai Y. A common plan for dorsoventral patterning in Bilateria. Nature. 1996;380:37–40. doi: 10.1038/380037a0. [DOI] [PubMed] [Google Scholar]
- 68.Herpin A, Lelong C, Favrel P. Transforming growth factor-beta-related proteins: an ancestral and widespread superfamily of cytokines in metazoans. Dev Comp Immunol. 2004;28:461–85. doi: 10.1016/j.dci.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 69.Kingsley DM. The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8:133–46. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 70.Cavaillon JM. The historical milestones in the understanding of leukocyte biology initiated by Elie Metchnikoff. J Leukoc Biol. 2011;90:413–24. doi: 10.1189/jlb.0211094. [DOI] [PubMed] [Google Scholar]
- 71.Metchnikoff E. Immunity in infective diseases. Johnson Reprint Corporation; New York and London: 1905. 1968; orig. [Google Scholar]
- 72.Smith LC, Ghosh J, Buckley KM, Clow LA, Dheilly NM, Haug T, Henson JH, Li C, Lun CM, Majeske AJ, Matranga V, Nair SV, Rast JP, Raftos DA, Roth M, Sacchi S, Schrankel CS, Stensvag K. Soderhall K, editor. Echinoderm immunity. Invertebrate Immunity. 2010;708:260–301. doi: 10.1007/978-1-4419-8059-5_14. [DOI] [PubMed] [Google Scholar]
- 73.West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–U543. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Underhill DM, Ozinsky A. Phagocytosis of microbes: complexity in action. Annu Rev Immunol. 2002;20:825–52. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 75.Danilova N. The evolution of immune mechanisms. J Exp Zool B Mol Dev Evol. 2006;306:496–520. doi: 10.1002/jez.b.21102. [DOI] [PubMed] [Google Scholar]
- 76.Jung HC, Eckmann L, Yang SK, Panja A, Fierer J, Morzyckawroblewska E, Kagnoff MF. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J Clin Invest. 1995;95:55–65. doi: 10.1172/JCI117676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maderna P, Godson C. Phagocytosis of apoptotic cells and the resolution of inflammation. Biochim Biophys Acta. 2003;1639:141–151. doi: 10.1016/j.bbadis.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 78.Henson PM, Bratton DL, Fadok VA. Apoptotic cell removal. Curr Biol. 2001;11:R795–R805. doi: 10.1016/s0960-9822(01)00474-2. [DOI] [PubMed] [Google Scholar]
- 79.Havixbeck JJ, Rieger AM, Wong ME, Wilkie MP, Barreda DR. Evolutionary conservation of divergent pro-inflammatory and homeostatic responses in Lamprey phagocytes. PLoS One. 2014;9:e86255. doi: 10.1371/journal.pone.0086255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Esmann L, Idel C, Sarkar A, Hellberg L, Behnen M, Moller S, van Zandbergen G, Klinger M, Kohl J, Bussmeyer U, Solbach W, Laskay T. Phagocytosis of apoptotic cells by neutrophil granulocytes: diminished proinflammatory neutrophil functions in the presence of apoptotic cells. J Immunol. 2010;184:391–400. doi: 10.4049/jimmunol.0900564. [DOI] [PubMed] [Google Scholar]
- 81.Devitt A, Marshall LJ. The innate immune system and the clearance of apoptotic cells. J Leukoc Biol. 2011;90:447–57. doi: 10.1189/jlb.0211095. [DOI] [PubMed] [Google Scholar]
- 82.Silva MT. Macrophage phagocytosis of neutrophils at inflammatory/infectious foci: a cooperative mechanism in the control of infection and infectious inflammation. J Leukoc Biol. 2011;89:675–683. doi: 10.1189/jlb.0910536. [DOI] [PubMed] [Google Scholar]
- 83.Soehnlein O, Lindbom L. Phagocyte partnership during the onset and resolution of inflammation. Nature Rev Immunol. 2010;10:427–439. doi: 10.1038/nri2779. [DOI] [PubMed] [Google Scholar]
- 84.Mulero V, Sepulcre MP, Rainger GE, Buckley CD. Editorial: Neutrophils live on a two-way street. J Leukoc Biol. 2011;89:645–7. doi: 10.1189/jlb.0111013. [DOI] [PubMed] [Google Scholar]
- 85.Scapini P, Cassatella MA. Social networking of human neutrophils within the immune system. Blood. 2014;124:710–719. doi: 10.1182/blood-2014-03-453217. [DOI] [PubMed] [Google Scholar]
- 86.Weissmann G. It's complicated: inflammation from Metchnikoff to Meryl Streep. FASEB J. 2010;24:4129–32. doi: 10.1096/fj.10-1101ufm. [DOI] [PubMed] [Google Scholar]
- 87.Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J Exp Med. 1973;137:1142–62. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jolles S. Paul Langerhans. J Clin Pathol. 2002;55:243. doi: 10.1136/jcp.55.4.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Langerhans P. Ueber die Nerven der menschlichen Haut Archiv für pathologische Anatomie und Physiologie und für klinische Medicin. 1868;44:325–337. [Google Scholar]
- 90.Karmaus PW, Chi H. Genetic dissection of dendritic cell homeostasis and function: lessons from cell type-specific gene ablation. Cell Mol Life Sci. 2014;71:1893–906. doi: 10.1007/s00018-013-1534-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Satpathy AT, Wu X, Albring JC, Murphy KM. Re(de)fining the dendritic cell lineage. Nat Immunol. 2012;13:1145–54. doi: 10.1038/ni.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hume DA. Macrophages as APC and the dendritic cell myth. J Immunol. 2008;181:5829–35. doi: 10.4049/jimmunol.181.9.5829. [DOI] [PubMed] [Google Scholar]
- 93.Flajnik MF. Re-evaluation of the immunological Big Bang. Curr Biol. 2014;24:R1060–5. doi: 10.1016/j.cub.2014.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Flajnik MF, Du Pasquier L. Evolution of innate and adaptive immunity: can we draw a line? Trends Immunol. 2004;25:640–4. doi: 10.1016/j.it.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 95.Rumfelt LL, McKinney EC, Taylor E, Flajnik MF. The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand J Immunol. 2002;56:130–48. doi: 10.1046/j.1365-3083.2002.01116.x. [DOI] [PubMed] [Google Scholar]
- 96.Torroba M, Chiba A, Vicente A, Varas A, Sacedon R, Jimenez E, Honma Y, Zapata AG. Macrophage-lymphocyte cell clusters in the hypothalamic ventricle of some elasmobranch fish: ultrastructural analysis and possible functional significance. Anat Rec. 1995;242:400–10. doi: 10.1002/ar.1092420312. [DOI] [PubMed] [Google Scholar]
- 97.Ohta Y, Landis E, Boulay T, Phillips RB, Collet B, Secombes CJ, Flajnik MF, Hansen JD. Homologs of CD83 from elasmobranch and teleost fish. J Immunol. 2004;173:4553–60. doi: 10.4049/jimmunol.173.7.4553. [DOI] [PubMed] [Google Scholar]
- 98.Lugo-Villarino G, Balla KM, Stachura DL, Banuelos K, Werneck MB, Traver D. Identification of dendritic antigen-presenting cells in the zebrafish. Proc Natl Acad Sci U S A. 2010;107:15850–5. doi: 10.1073/pnas.1000494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wittamer V, Bertrand JY, Gutschow PW, Traver D. Characterization of the mononuclear phagocyte system in zebrafish. Blood. 2011;117:7126–35. doi: 10.1182/blood-2010-11-321448. [DOI] [PubMed] [Google Scholar]
- 100.Haugland GT, Jordal AE, Wergeland HI. Characterization of small, mononuclear blood cells from salmon having high phagocytic capacity and ability to differentiate into dendritic like cells. PLoS One. 2012;7:e49260. doi: 10.1371/journal.pone.0049260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bassity E, Clark TG. Functional identification of dendritic cells in the teleost model, rainbow trout (Oncorhynchus mykiss). PLoS One. 2012;7:e33196. doi: 10.1371/journal.pone.0033196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baldwin WM, Cohen N. A giant cell with dendritic cell properties in spleens of the anuran amphibian Xenopus laevis. Dev Comp Immunol. 1981;5:461–73. doi: 10.1016/s0145-305x(81)80058-4. [DOI] [PubMed] [Google Scholar]
- 103.Horton JD, Manning MJ. Effect of early thymectomy on the cellular changes occuring in the spleen of the clawed toad following administration of soluble antigen. Immunology. 1974;26:797–807. [PMC free article] [PubMed] [Google Scholar]
- 104.Aguzzi A, Kranich J, Krautler NJ. Follicular dendritic cells: origin, phenotype, and function in health and disease. Trends Immunol. 2014;35:105–13. doi: 10.1016/j.it.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 105.Krautler NJ, Kana V, Kranich J, Tian Y, Perera D, Lemm D, Schwarz P, Armulik A, Browning JL, Tallquist M, Buch T, Oliveira-Martins JB, Zhu C, Hermann M, Wagner U, Brink R, Heikenwalder M, Aguzzi A. Follicular dendritic cells emerge from ubiquitous perivascular precursors. Cell. 2012;150:194–206. doi: 10.1016/j.cell.2012.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Du Pasquier L, Flajnik MF. Expression of MHC class II antigens during Xenopus development. Dev Immunol. 1990;1:85–95. doi: 10.1155/1990/67913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Turpen JB, Smith PB. Analysis of hemopoietic lineage of accessory cells in the developing thymus of Xenopus laevis. J Immunol. 1986;136:412–21. [PubMed] [Google Scholar]
- 108.Mescher AL, Wolf WL, Moseman EA, Hartman B, Harrison C, Nguyen E, Neff AW. Cells of cutaneous immunity in Xenopus: studies during larval development and limb regeneration. Dev Comp Immunol. 2007;31:383–93. doi: 10.1016/j.dci.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 109.Garcia Barrutia MS, Leceta J, Fonfria J, Garrido E, Zapata A. Non-lymphoid cells of the anuran spleen: an ultrastructural study in the natterjack, Bufo calamita. Am J Anat. 1983;167:83–94. doi: 10.1002/aja.1001670108. [DOI] [PubMed] [Google Scholar]
- 110.Garcia Barrutia MS, Villena A, Gomariz RP, Razquin B, Zapata A. Ultrastructural changes in the spleen of the natterjack, Bufo calamita, after antigenic stimulation. Cell Tissue Res. 1985;239:435–41. doi: 10.1007/BF00218024. [DOI] [PubMed] [Google Scholar]
- 111.Kroese FG, Leceta J, Dopp EA, Herraez MP, Nieuwenhuis P, Zapata A. Dendritic immune complex trapping cells in the spleen of the snake, Python reticulatus. Dev Comp Immunol. 1985;9:641–52. doi: 10.1016/0145-305x(85)90029-1. [DOI] [PubMed] [Google Scholar]
- 112.Zapata A, Leceta J, Barrutia MG. Ultrastructure of splenic white pulp of the turtle, Mauremys caspica. Cell Tissue Res. 1981;220:845–55. doi: 10.1007/BF00210466. [DOI] [PubMed] [Google Scholar]
- 113.Leceta J, Zapata AG. White pulp compartments in the spleen of the turtle Mauremys caspica - a light-microscopic, electron-microscopic, and immunohistochemical study. Cell Tissue Res. 1991;266:605–613. [Google Scholar]
- 114.Olah I, Nagy N. Retrospection to discovery of bursal function and recognition of avian dendritic cells; past and present. Dev Comp Immunol. 2013;41:310–5. doi: 10.1016/j.dci.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 115.Wu Z, Rothwell L, Young JR, Kaufman J, Butter C, Kaiser P. Generation and characterization of chicken bone marrow-derived dendritic cells. Immunology. 2010;129:133–45. doi: 10.1111/j.1365-2567.2009.03129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu Z, Hu T, Kaiser P. Chicken CCR6 and CCR7 are markers for immature and mature dendritic cells respectively. Dev Comp Immunol. 2011;35:563–7. doi: 10.1016/j.dci.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 117.Del Cacho E, Gallego M, Lillehoj HS, Lopez-Bernard F, Sanchez-Acedo C. Avian follicular and interdigitating dendritic cells: isolation and morphologic, phenotypic, and functional analyses. Vet Immunol Immunopathol. 2009;129:66–75. doi: 10.1016/j.vetimm.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 118.Kaiser P. The long view: a bright past, a brighter future? Forty years of chicken immunology pre- and post-genome. Avian Pathol. 2012;41:511–8. doi: 10.1080/03079457.2012.735359. [DOI] [PubMed] [Google Scholar]
- 119.Magor KE, Miranzo Navarro D, Barber MR, Petkau K, Fleming-Canepa X, Blyth GA, Blaine AH. Defense genes missing from the flight division. Dev Comp Immunol. 2013;41:377–88. doi: 10.1016/j.dci.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]