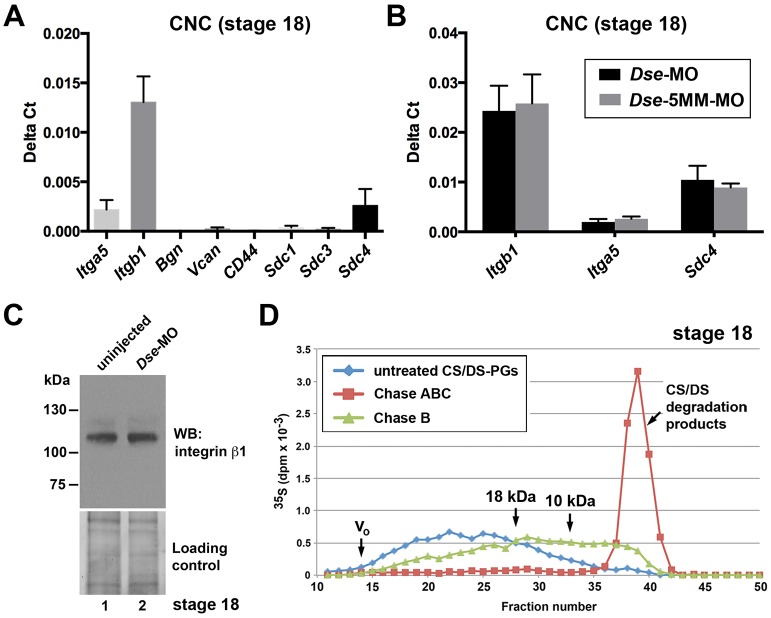
Fig. 7.
CS/DS-PGs in CNC cells. (A) qPCR analysis in uninjected CNC explants at stage 18. Note abundant expression of Itga5, Itgb1 and Sdc4. Results are mean±s.d. from triplicates (n≥4 biological replicates). (B) Dse-MO does not differentially affect the mRNA levels of Itga5, Itgb1 and Sdc4 compared with Dse-5MM-MO. Results are mean±s.d. (n≥4 biological replicates). (C) Dse-MO does not reduce the protein amount of integrin β1 in explants enriched in neural crest and epidermis of stage 18 embryos. Western blotting was performed on a 7.5% Mini-Protean TGX Stain-free gel (Bio-Rad). The loading control was ascertained prior to blotting using the ChemiDoc Touch Imaging System. Resuts is representative of two independent experiments (n=2). (D) Metabolic labeling of PGs in stage 18 CNC explants. Note that Chase B partially degrades CS/DS PGs >18 kDa. The IdoA is a rare modification because the split chains are ∼10 kDa.