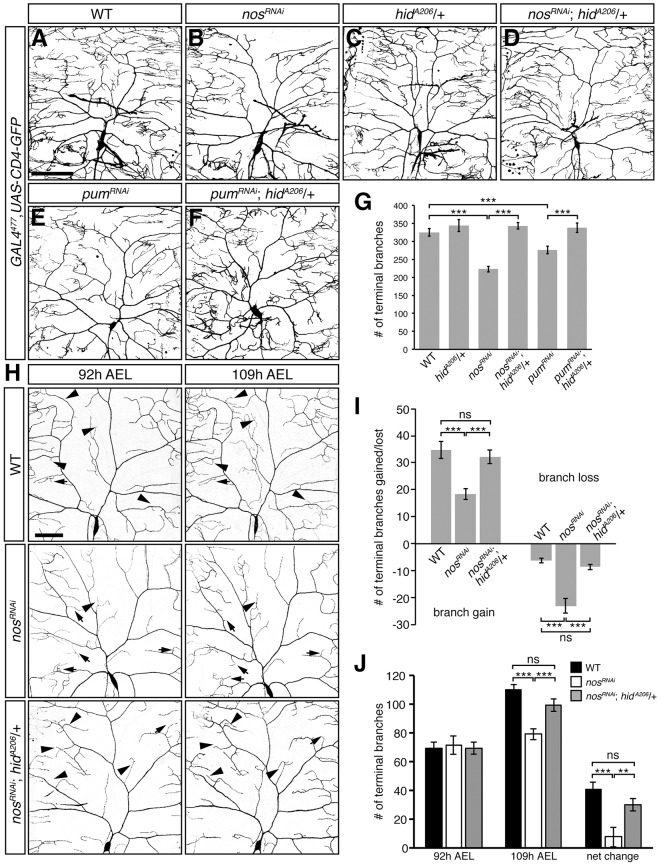
Fig. 1.
Reducing hid function rescues dendritic arborization defects of class IV da neurons lacking nos and pum. (A-F) Confocal z-series projections of representative class IV da neurons from wild-type (WT) (A), nosRNAi (B), hidA206/+ (C), nosRNAi; hidA206/+ (D), pumRNAi (E) and pumRNAi; hidA206/+ (F) larvae. (G) Quantification of the total number of terminal branches. (H) Class IV da neurons in WT, nosRNAi and nosRNAi; hidA206/+ larvae imaged at 92 h and 109 h AEL. Arrowheads indicate examples of new branch outgrowth or branch elongation and arrows designate examples of branch retraction/or loss. In A-F and H, GAL4477 was used to drive expression of UAS-RNAi transgenes and UAS-CD4-GFP (to mark the neurons). (I) Quantification of the number of new and/or elongating branches and lost or retracting branches between time points. Net change represents the difference in the average number of terminal branches observed between 92 h and 109 h AEL class IV da neurons. (J) Quantification of the number of dendritic termini in WT, nosRNAi and nosRNAi; hidA206/+ larvae at 92 h AEL and 109 h AEL. Values in G,I-J are mean±s.e.m., n≥10 for each genotype; ns, not significant; **P≤0.01, ***P≤0.001 by two-tailed Student's t-test. Scale bars: 100 μm.