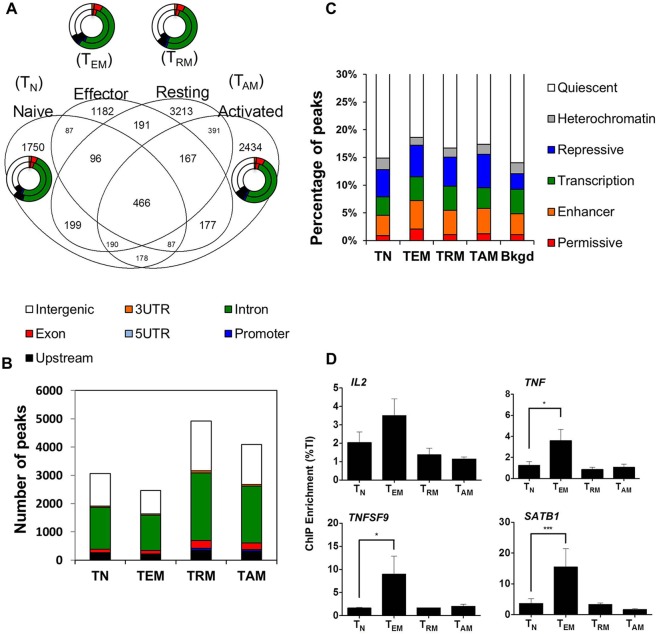
Fig. 2.
Distribution of chromatinized PKC-θ in memory CD4+ T cells. (A) Overlapping PKC-θ-bound regions in ex-vivo-derived naïve (TN), effector (TEM), resting (TRM) and activated memory (TAM) CD4+ T cells. Graphs show genomic annotation of unique peaks (outside ring) compared to the peaks common to all subsets (inside ring). (B) PKC-θ peaks in memory CD4+ T cell subsets categorized into intergenic, 3′ untranslated region (UTR), intron, exon, 5′ UTR, promoter and upstream regions according to the nearest ENSEMBL gene, see corresponding colors in A. (C) The percentage of PKC-θ peaks in memory CD4+ T cell subsets and genomic background (Bkgd) annotated according to their chromatin state segmentation: permissive (H3K4me3, H3K27ac and H2K9ac), transcription (H3K36me3), enhancer (H3K4me1, and to different degrees H3K27ac or H3K9ac), repressed (H3K27me3 and/or H3K9me3), heterochromatin (H3K9me3) and quiescent (associated with low levels of histone marks). (D) Chromatin immunoprecipitation (ChIP-PCR) analysis on PKC-θ at the proximal promoters of the transcriptional-memory-responsive genes: IL2, TNF and TNFSF9 and an intronic region of SATB1. ChIP enrichment is shown as a percentage relative to the total input (TI) with background subtraction (mean±s.e.m., n=3). *P≤0.05, ***P≤0.001 (one-tailed Student's t-test).