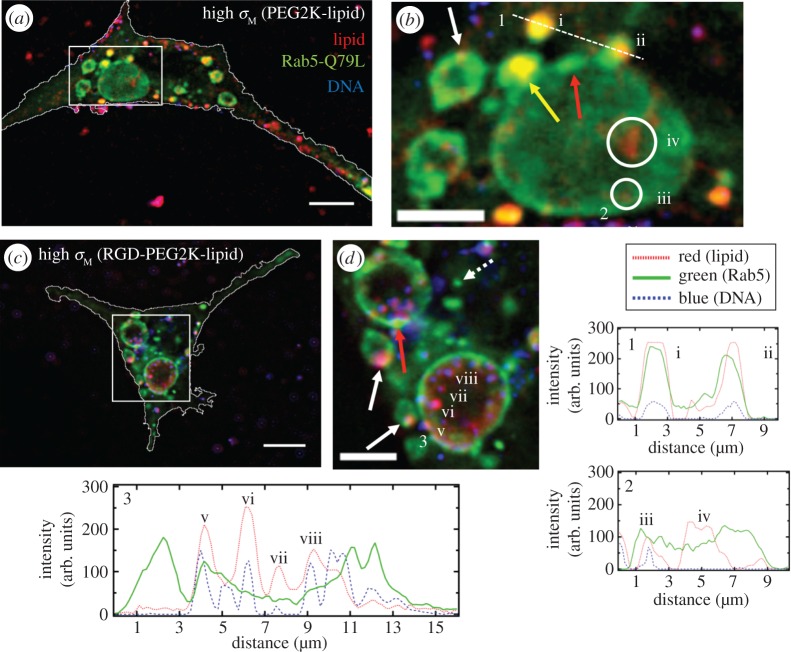
Figure 6.
Colocalization of cationic liposome–DNA nanoparticles (CL–DNA NPs) and giant early endosomes (GEEs). Rab5-Q79L inhibits endosome maturation, giving rise to large (greater than 5 μm) endosomes with spatially resolvable NPs. Nearly all intracellular CL–DNA NPs are foundwithin Rab5-Q79L-GFP labelled endosomes (red, TRITC (lipid); green, GFP (endosomes); blue, Cy5 (DNA)). All cells shown were fixed after 1 h of incubation at 4°C followed by 1 h of incubation at 37°C. (a,c) Fluorescent micrographs and cell boundaries (white outlines) of L-cells with (a) PEGylated high-σM NPs (MVL5/DOPC/PEG2K-lipid at a molar ratio of 50/40/10), (c) RGD-tagged high-σM NPs (MVL5/DOPC/RGD-PEG2K-lipid at a molar ratio 50/45/5). (b,d) High magnification of boxed regions in (a,c). (1–3) Intensity profiles of labelled scans from high magnification regions. Intensity profiles 1 and 2 show NPs (i,ii,iii) and liposomes (iv) found in EEs (i, ii) and GEEs (iii).Intensity profile 3 shows clear evidence of individually resolvable NPs (v,vi,vii,viii) inside the lumen of the GEE including a NP interacting with the inner membrane of the GEE (v). The yellow arrow in (b) points to an EE containing a NP fusing with a GEE. Red arrows in (b,d) show GFP-rich regions of the GEE membrane. Dashed arrow in (d) shows a smaller EE, similar to what is observed with wild-type Rab5. Solid white arrows in (b,d) point to clear examples of NPs adhering to the GEE membrane. Scale bars in (a,c) and (b,d) are 10 μm and 5 μm, respectively. Adapted from [63] with permission from Elsevier.