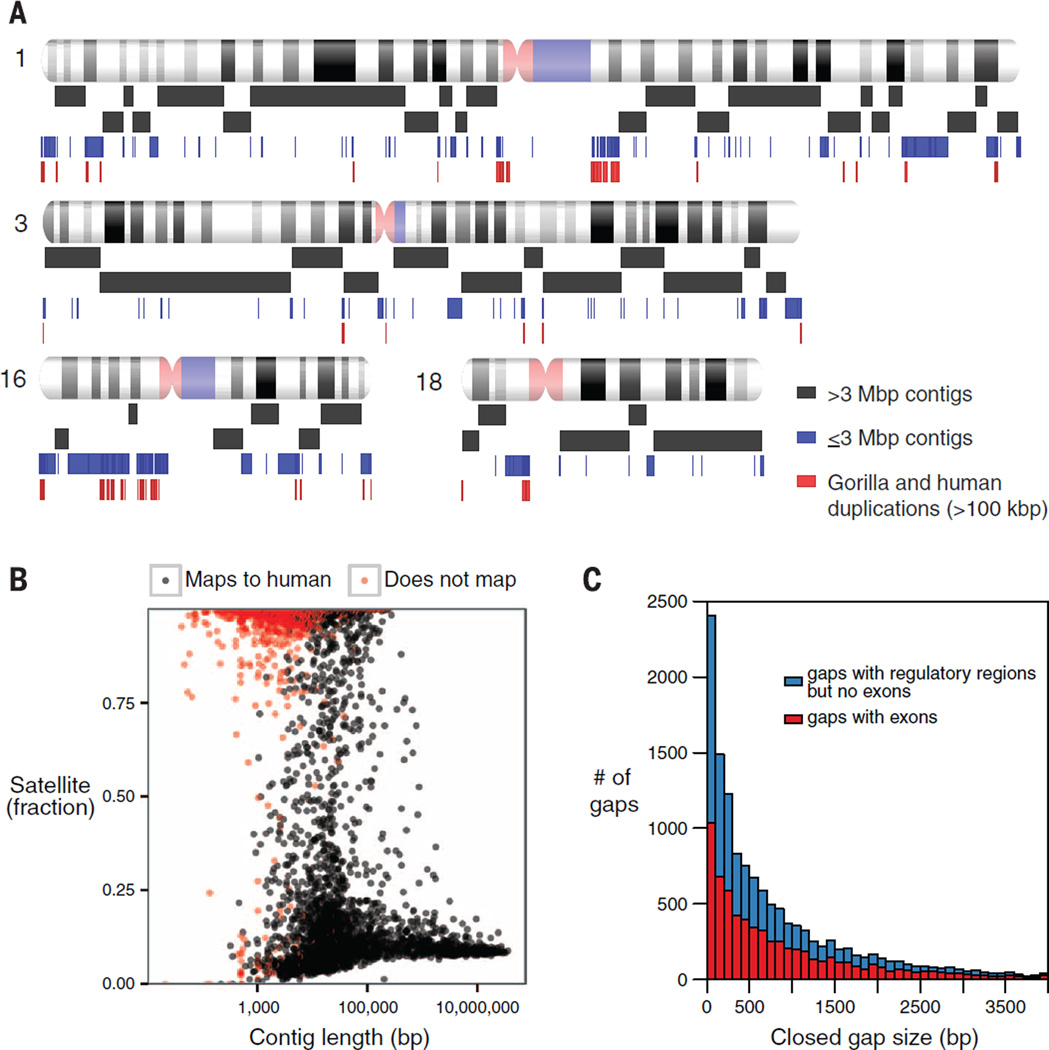
Fig. 1. Gorilla genome assembly.
(A) Schematic depicting assembly contig lengths (contig N50 = 9.6 Mbp) mapped to human GRCh38 chromosomes. The first two rows of black rectangles represent contigs >3 Mbp, the blue rectangles correspond to contigs ≤3 Mbp, and red rectangles correspond to blocks of human/gorilla segmental duplications >100 kbp. (B) Mappability and satellite content of Susie3 contigs. Satellite content defined by use of RepeatMasker (28) and Tandem Repeats Finder (29). Contigs that are unable to map to GRCh38 by using BLASR (colored red) (30) contain a high fraction of satellite sequence. (C) Length distribution of gaps in the published gorilla assembly gorGor3 closed by Susie3 and containing exons or regulatory regions. Of the gaps in gorGor3, 94% were closed in Susie3, with thousands corresponding to missing exons (red) and putative noncoding regulatory DNA (blue).