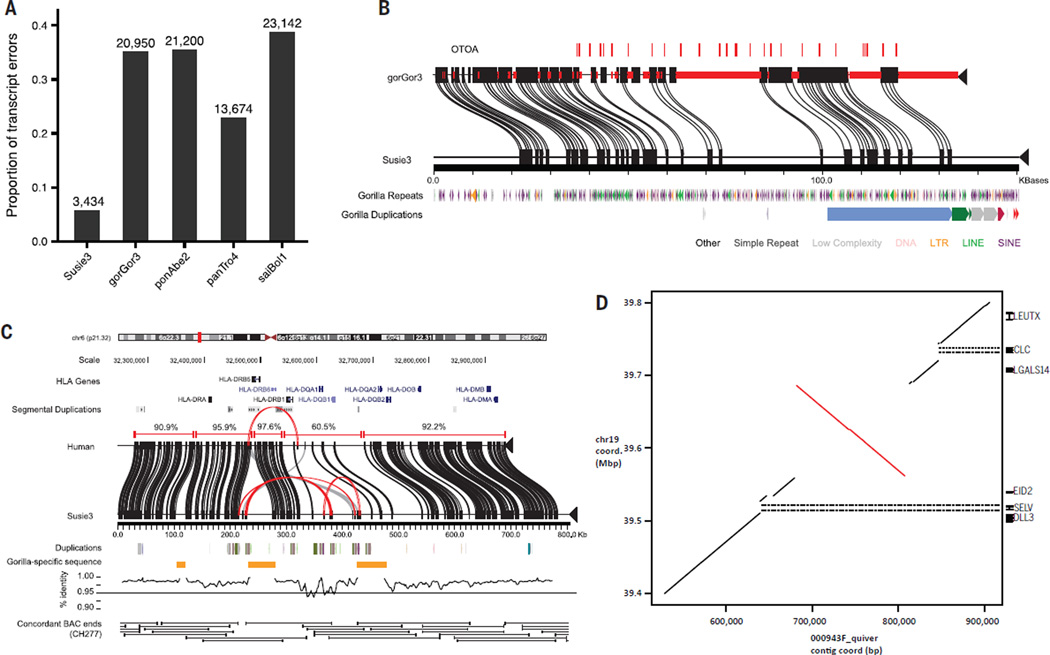
Fig. 4. Gene annotation and structural variation.
(A) Proportion of GENCODE transcripts with assembly errors when aligned with gorilla assemblies Susie3 and gorGor3, and three reference assemblies, including orangutan (ponAbe2), chimpanzee (panTro4), and squirrel monkey (saiBol1). Examples of assembly errors include transcript mappings extending off the end of contigs/scaffolds, containing unknown bases, or incomplete transcript mapping. (B) An example of a gene, otoancorin (OTOA), with complete exon representation (red ticks) resolved in the new assembly. Red bars on gorGor3 sequence indicate gaps in the assembly. Alignments between gorilla assemblies are based on Miropeats (31). (C) Alignment of MHC Class II locus in Susie3 against GRCh37 with Miropeats. Alignment identities of collinear blocks between assemblies are shown above the corresponding GRCh37 sequence. Repeats internal to Susie3 are shown in red along the coordinates. Alignment identity across the entire locus is shown below the Susie3 contigs in 5-kbp windows (1 kbp sliding). Support for the proper organization of the Susie3 sequence is shown by the tiling path of concordant BAC end sequences from the Kamilah BAC library (CHORI-277). (D) A sequence-resolved complex gorilla genome structural variation orthologous to human chromosome 19:38,867,213–39,866,620 (GRCh38). The dot-matrix plot shows a 125,375-bp inversion flanked by a proximal 16-kbp deletion and 8-kbp insertion, and a 23-kbp distal deletion. The deletions remove the entire sequences of the SELV and CLC genes in gorilla when compared with human.