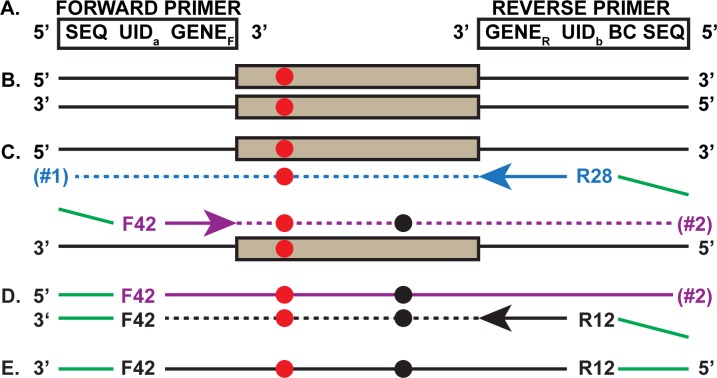
Fig 1. First PCR Round of the SSS strategy.
a. Two primers (forward and reverse) are used for the first two SSS PCR cycles (all primers are listed in S1 Fig). Each primer (5’ to 3’) contains a Sequencing primer region (SEQ), a Unique Identifier (UID) and the forward (GENEF) or reverse (GENER) versions of the gene specific DNA sequences. Only the reverse primer contains a barcode for multiplex analysis (BC). b. Duplex genomic DNA fragment carrying a gene target that experienced an in vivo germline mutation (red circle). c. The two new daughter strands of the first denaturation, primer hybridization and primer extension. The F42 and R28 represent one of the many UIDs on the forward and reverse primers, respectively. The blue arrow indicates the direction of the reverse primer extension that includes the specific target region (blue dashes) and extends through the target as does the purple forward primer on the other strand (purple dashes). The green overhang at the extreme 5’ ends will anneal to the primers used in the second PCR round. Two different extension products are created (#1 and #2). Notice that extension product #2, with the already existing in vivo mutation is (for convenience only) also shown to experience a new in vitro mutation (black circle). In the subsequent steps, for simplicity, we only follow one of the two extension products (#2). d. The second cycle with the same primer mix, invariably involves a primer (black) with a different UID (R12) and copies DNA strand #2 (black dashes). Subsequent single strand-specific exonuclease digestion removes all the initial PCR primers, trims the 3’ DNA overhangs from the second cycle primer extension products and any remaining single-stranded genomic DNA. e. Importantly, F42-R12 is tagged by different UIDs at each end and is then subjected to the second round of PCR (see Fig 2).