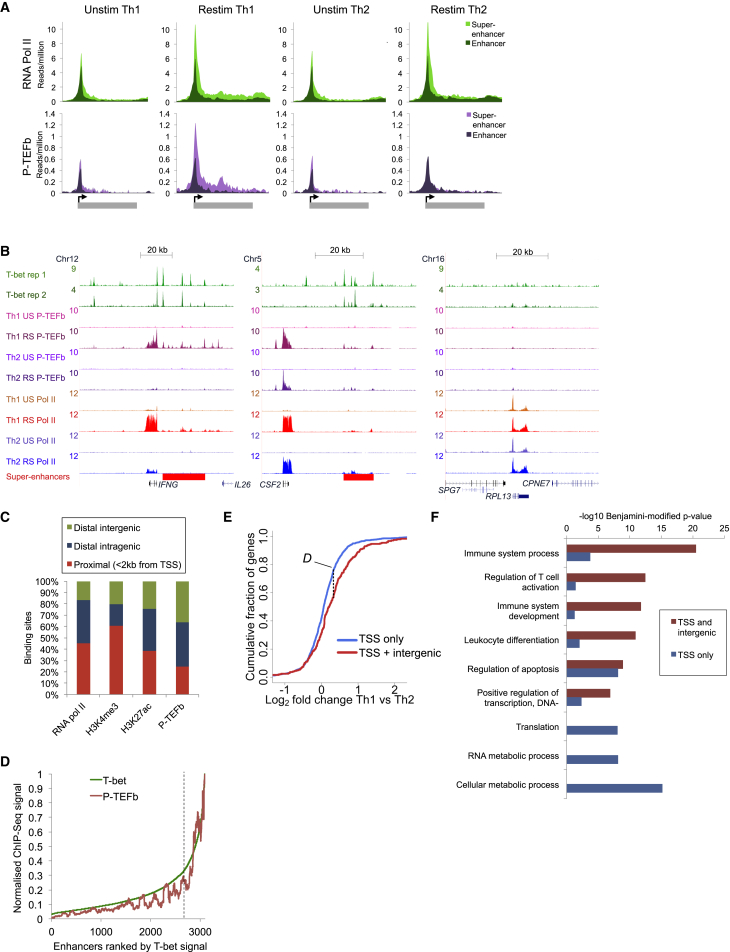
Figure 2.
Extensive P-TEFb Binding at Super-Enhancers and Associated Genes
(A) Average number of ChIP-seq reads for RNA pol II and P-TEFb in unstimulated (US) or restimulated (RS) human Th1 and Th2 cells. All genes are bound by RNA pol II in at least one condition and divided into those associated with a super-enhancer (n = 231) or a typical enhancer (n = 1,307).
(B) T-bet, P-TEFb, and RNA pol II binding at IFNG, CSF2 (associated with T-bet super-enhancers), and the housekeeping gene RPL13.
(C) Percentage of sites for RNA pol II, H3K4me3, H3K27ac, and P-TEFb in Th1 cells that are proximal (<2 kb from TSS), distal intragenic, or distal intergenic.
(D) Distribution of T-bet and P-TEFb ChIP-seq signals across 3,191 T-bet Th1 enhancers, ranked according to T-bet signal. The enhancers classified as T-bet super-enhancers are to the right of the vertical dashed line. The ChIP signals are shown as moving averages (window size of 100 bp).
(E) Cumulative distribution frequency of gene expression in Th1 cells relative to Th2 cells for genes occupied by P-TEFb at the gene TSS only (blue, n = 970) or at the TSS and an intergenic site (red, n = 245) (D = 0.19, p = 7.47 × 10−7 [K-S test]).
(F) Significance of the enrichment of biological process gene ontology categories in the set of genes occupied by P-TEFb at the gene TSS only (blue) or at the gene TSS and an intergenic site (red).
See also Figure S2.