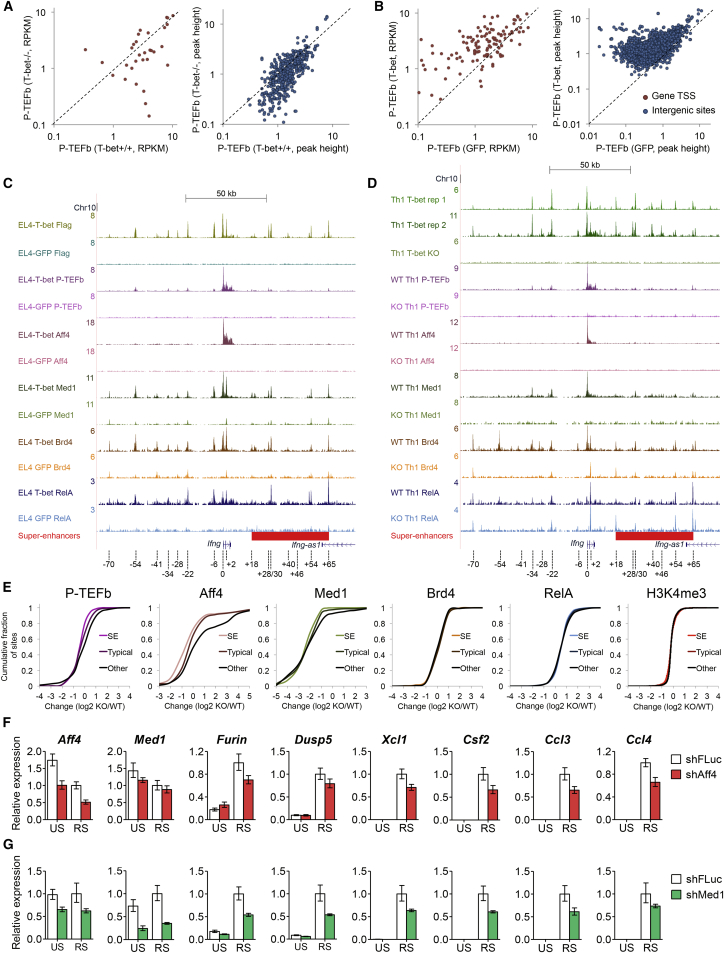
Figure 4.
T-bet Is Necessary for Recruitment of Mediator, the SEC, and P-TEFb to Genes and Super-Enhancers
(A) Scatterplot showing the read density (RPKM) for P-TEFb at super-enhancer-associated genes occupied by P-TEFb in primary mouse T-bet+/+ versus T-bet−/− Th1 cells (left). A scatterplot showing the P-TEFb peak height (reads/million) at intergenic T-bet binding sites (p < 10−9) in primary mouse T-bet+/+ versus T-bet−/− Th1 cells is on the right.
(B) As in (A), except for activated EL4 cells stably expressing GFP (x axis) versus cells stably expressing T-bet and GFP (y axis).
(C) ChIP-seq binding profiles for T-bet, P-TEFb, RelA, Brd4, Med1, and Aff4 in EL4 cells stably expressing GFP alone or EL4 cells stably expressing FLAG-T-bet and GFP, with both cell lines restimulated with PMA and ionomycin. The positions of transcription factor binding sites relative to the Ifng TSS are marked (to the nearest kb).
(D) As in (C), except for WT and T-bet−/− Th1 cells restimulated with PMA and ionomycin.
(E) Cumulative distribution frequency of the change in P-TEFb, Aff4, Med1, Brd4, RelA, and H3K4me3 occupancy between T-bet−/− and T-bet+/+ cells at super-enhancers and associated genes (SE), at typical enhancers and associated genes (Typical), and at other sites (Other).
(F) Expression of Aff4 and Med1 and the T-bet target genes Furin, Dusp5, Xcl1, Csf2, Ccl3, and Ccl4 relative to Hprt (mean and SD, n = 2 biological replicates) in unstimulated and restimulated Th1 cells transduced with retroviruses encoding shRNAs against luciferase (white) or Aff4 (red).
(G) As in (F), except for shRNAs to luciferase (white) or Med1 (green) (mean and SD, n = 3 technical replicates). A replicate experiment with combined shRNA knock down of Med1 and Med17 is shown in Figure S4.