Figure 6. PCR does not bias SHAPE-Seq reactivity calculations.
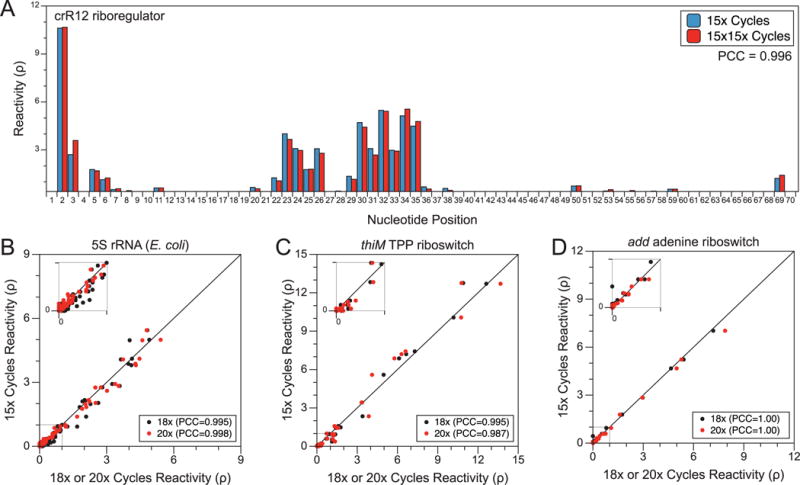
(A) Comparison of the crR12 riboregulator calculated reactivities from an in-cell SHAPE-Seq experiment [26] using either 15× cycles of PCR (15x; blue) vs. 15× cycles without PE_F, followed by another 15× cycles including PE_F (15×15×; red). A Pearson Correlation Coefficient (PCC) value of 0.996 suggests increased PCR cycling does not affect the reactivity calculation. (B) Base-wise comparison of reactivities calculated from 5S rRNA SHAPE-Seq libraries using 15× cycles of PCR vs. 18× (black) or 20× (red) cycles of PCR (RMDB: 5SRRNA_1M7_0009). A PCC near unity suggests little difference in the reactivity values calculated from libraries with increased PCR cycling. Similar analyses were done for the thiM TPP aptamer domain (RMDB: TPPSC_1M7_0005) (C) and add adenine aptamer domain (D) (RMDB: ADDSC_1M7_0008).
