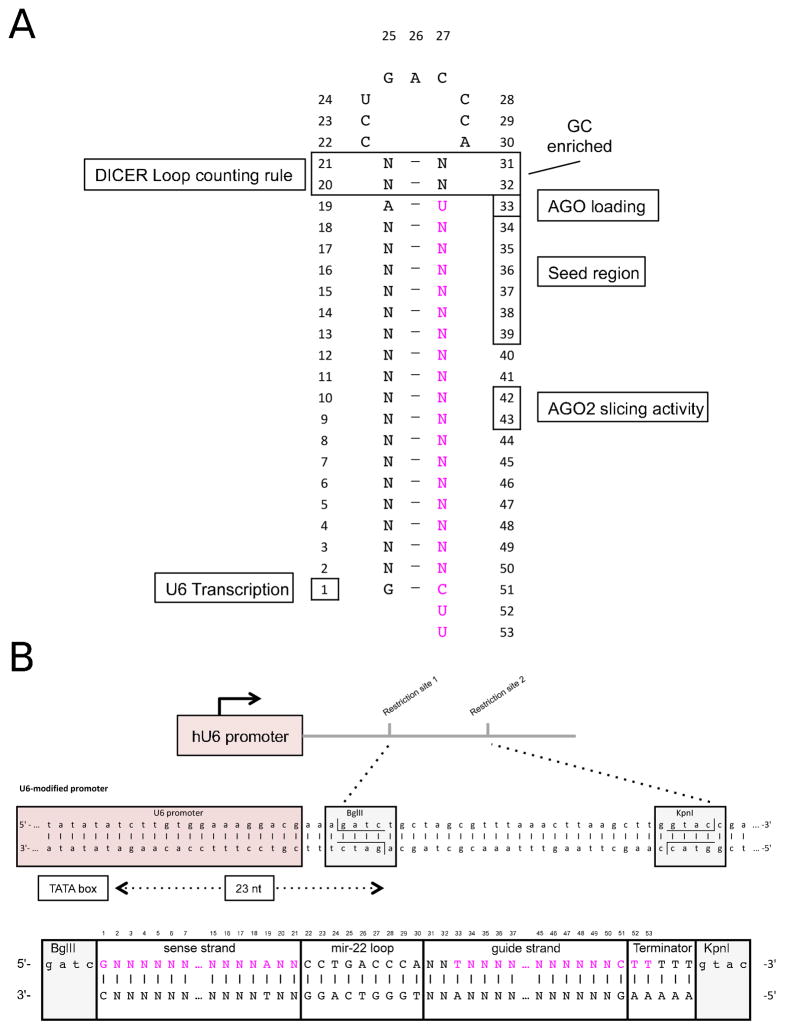
Fig 4. Cloning design for optimal processing of pre-miRNA-like shRNA.
A) shRNA hairpin structure containing a detailed memorandum of the reported sequence and structural features involved in the correct processing by Dicer and loading into AGO proteins. B) Expression though U6 promoter (Pol III) requires a precise start and termination of the transcription of the shRNA hairpin. To achieve this precision on the starting of the transcription, the first nucleotide of the shRNA should be located 23 nt from the TATA box. In this scheme we show the precise location of a BglII restriction site introduced to facilitate this precise cloning. Below, we present an oligonucleotide design containing the passenger sequence, the loop of mir-22, the guide sequence of the shRNA, a poly(T) terminator, and flanked by the required BglII and KpnI overhangs.