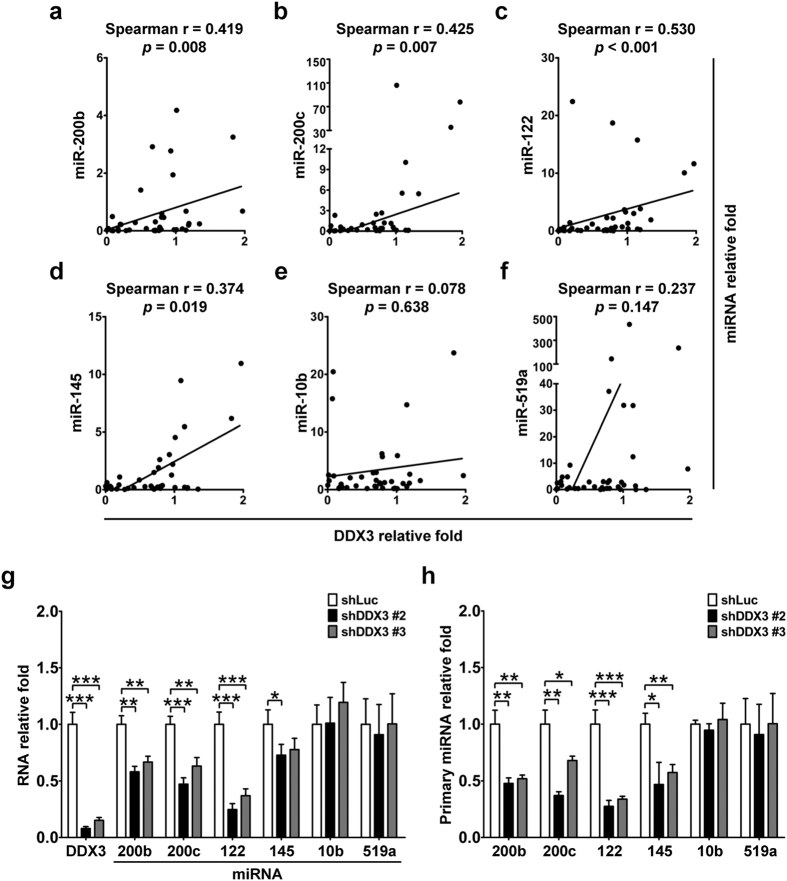
Figure 5. DDX3 level correlates with expression of several tumor-suppressive miRNAs.
Expressions of miRNAs and U6 snRNA as well as mRNA transcripts of DDX3 and GAPDH in 39 paired tumor and non-tumor tissues of HCC patients from TLCN were analyzed by qRT-PCR. U6 snRNA and GAPDH transcripts were used as internal control for miRNA and DDX3 expression, respectively. Fold change of each transcript in tumor tissues was relative to that of corresponding non-tumor tissues. Correlation of (a) miR-200b, (b) miR-200c, (c) miR-122, (d) miR-145, (e) miR-10b and (f) miR-519a expression with DDX3 level was performed by Spearman correlation criteria. p < 0.05 represents statistical significance. (g) Knockdown of DDX3 reduced expressions of tumor-suppressive miRNAs. Total RNA extracted from shLuc, shDDX3 #2 and shDDX3 #3 cells was subjected to qRT-PCR for detection of miR-200b, miR-200c, miR-122, miR-145, miR-10b, miR-519a and U6 snRNA as well as DDX3 and GAPDH transcripts. U6 snRNA and GAPDH transcripts were served as internal control for miRNA and DDX3 expression, respectively. Fold change of each transcript in shDDX3 #2 and shDDX3 #3 cells was relative to that of shLuc cells. Experiments were performed at least three times, and the error bar indicates ± 1 s.d. of the mean. Statistical analyses were carried out using t test (*p < 0.05; **p < 0.01; ***p < 0.001). (h) DDX3 knockdown repressed primary transcript expressions of tumor-suppressive miRNAs. Primary transcript expressions of miR-200b, miR-200c, miR-122, miR-145, miR-10b, miR-519a and GAPDH in shLuc, shDDX3 #2 and shDDX3 #3 cells were analyzed by qRT-PCR. GAPDH was used as internal control. Fold change of each transcript in shDDX3 #2 and shDDX3 #3 cells was relative to that of shLuc cells. Results were derived from at least three independent experiments, and the error bar indicated ± 1 s.d. of the mean. Statistical analyses were carried out using t test (*p < 0.05; **p < 0.01; ***p < 0.001).