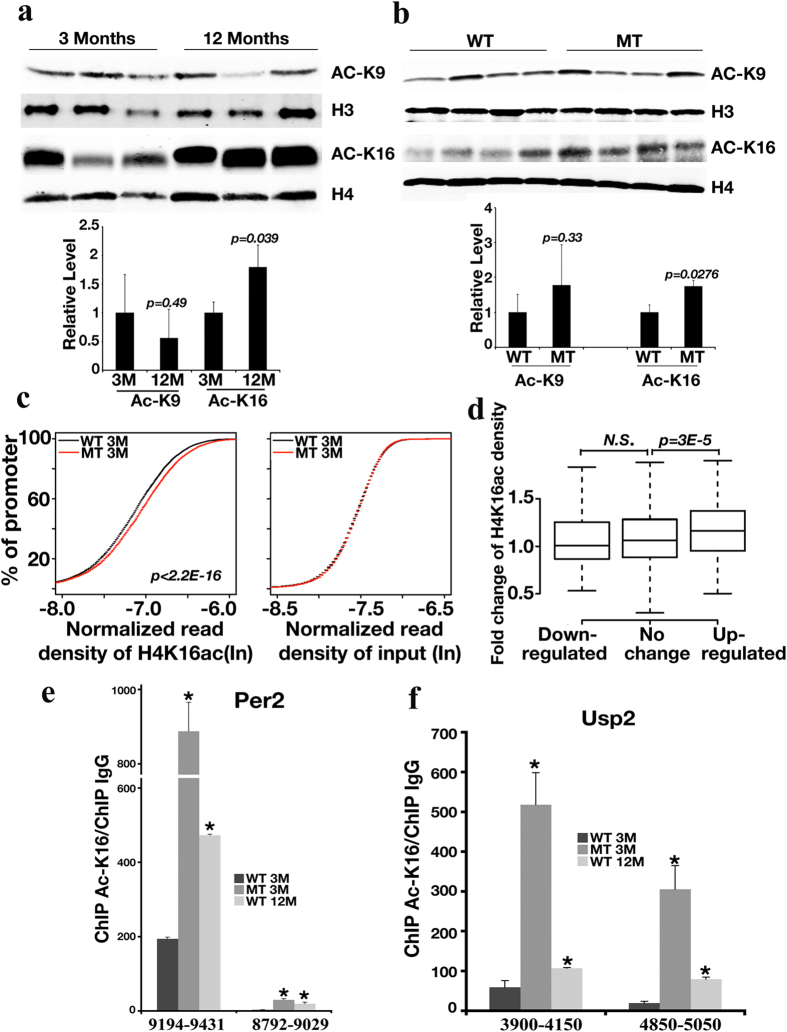
Figure 3. Sirt1 Deletion Induces Global Increase of Ac-K16 of Histone H4.
(a,b) Western blot analysis of histone H4K16 and H3K9 acetylation level in 3- and 12-month-old WT mice (n = 3) (a) and 3-month-old WT and MT mice (n = 4) (b). Upper panel is the representative image of western blot, the lower panel is the quantification of western blots. (c) Cumulative distribution of ChIP-Seq read density for H4K16ac at gene promoters for 3-month-old WT and MT mice. Livers from 2 pairs of mice were utilized for ChIP-Seq analysis. Y-axis shows the percentage of promoters that exhibit H4K16ac. A line shifted to the left means a systematically lower level of H4K16ac. p-value was calculated by the Kolmogorov-Smirnov test. The analysis was repeated by using input read densities for comparison purposes. (d) Boxplot of the fold-change of normalized H4K16ac densities between 3-month-old WT and MT mice at the promoter regions of genes up-regulated (150 genes), down-regulated (165 genes), and not changed (8937 genes) in the 3-month-old MT mice compared with the 3-month-old WT mice. The top and bottom of the box, and the band near the middle of the box represent 75th, 25th, and 50th percentiles, respectively. N.S., not significant. (e,f) Validation of H4K16ac enrichment in promoters of Sirt1-dependent aging-related up-regulated genes by qChIP. Data represent the mean (±SD, n = 3) *p < 0.05 when compared with WT 3-month-old samples. M, months; WT, wild-type; MT, mutant (Sirt1-deficient). qChIP data were first normalized with individual input, then normalized again with qChIP of IgG. Primer positions are shown in supplementary Figure 2.