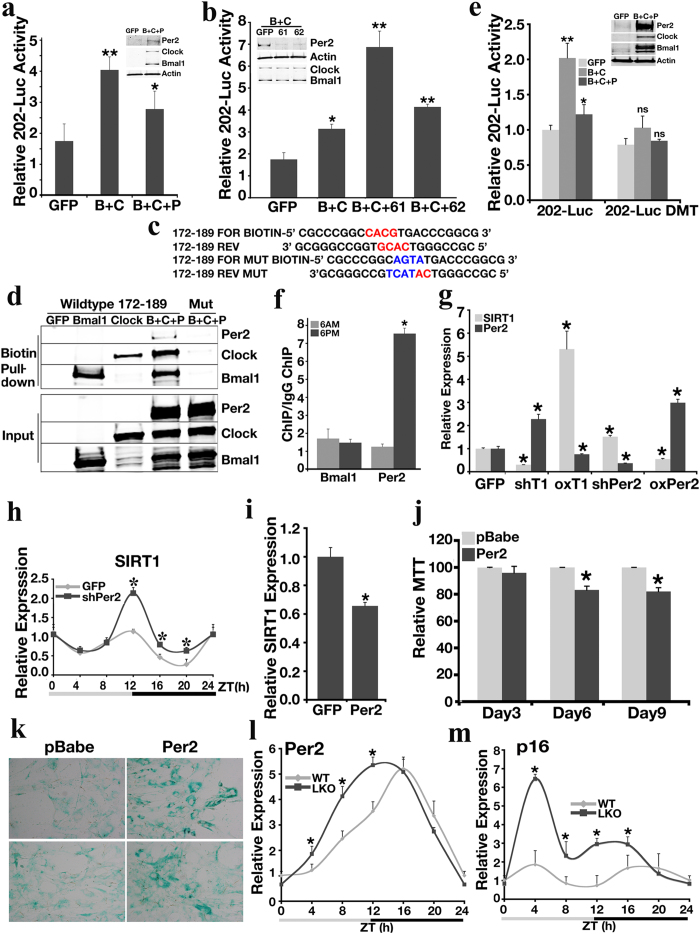
Figure 5. Sirt1 and Per2 Constitute a Reciprocal Negative Regulation Loop in vitro and in vivo.
(a) Sirt1 promoter luciferase reporter activity of 202-Luc construct following ectopic expression of Bmal1 (b), Clock (c), and Per2 (P) in MEFs. (b) Effect of Per2 knockdown on Sirt1 promoter 202-Luc construct luciferase reporter activity in response to Bmal1 (b) and Clock (c) overexpression in MEFs. 61 and 62 are two independent shRNAs against Per2. (c) Biotin-labeled E-box sequences showing a possible Clock/Bmal1 binding site −(172–189). (d) Biotin pull-down assay examining Bmal1/Clock/Per2 binding to the Sirt1 promoter at E-box region −(172–189). (e) Sirt1 promoter 202-Luc luciferase reporter activity after mutation of the 2 E-boxes. DMT, mutant of two E-boxes core domains. The level of luciferase activity in GFP with WT 202-Luc cotransfected MEFs was set as 1. (f) ChIP analysis of Bmal1 and Per2 binding to the Sirt1 promoter in the liver of 3-month-old WT mice collected at ZT0 and ZT12. *p < 0.05 when compared with ZT0 data. (g) qRT-PCR analysis of SIRT1 and PER2 expression in primary human hepatocytes (n = 2) subjected to shRNA knockdown of SIRT1 (shT1) or PER2 (shPer2) or overexpression of SIRT1 (oxT1) or PER2 (oxPer2). The level of expression for GFP vector-transfected cells was set as 1. (h) Hepatic Sirt1 expression analyzed by qRT-PCR after 5 days of shPer2 virus infection. The level of expression of GFP-infected mice at ZT0 was set as 1. (i) Sirt1 expression in MEF cells infected with retroviruses containing either Per2 or pBabe analyzed by qRT-PCR. (j) MTT assay in MEF cells infected with Per2 or pBabe retrovirus. (k) β-galactosidase activity assay in MEF cells infected with Per2 or pBabe retrovirus. (l,m) qRT-PCR analysis of Per2 (l) and p16 (m) expression in the livers of 3-month-old WT and Sirt1 liver specific knockout (LKO) mice obtained by circadian liver sample collection. Data represent the mean (±SD). Either 3 mice or MEFs were utilized for each experiment, and the experiment was repeated 3 times in a triplicated fashion. **p < 0.01, *p < 0.05.