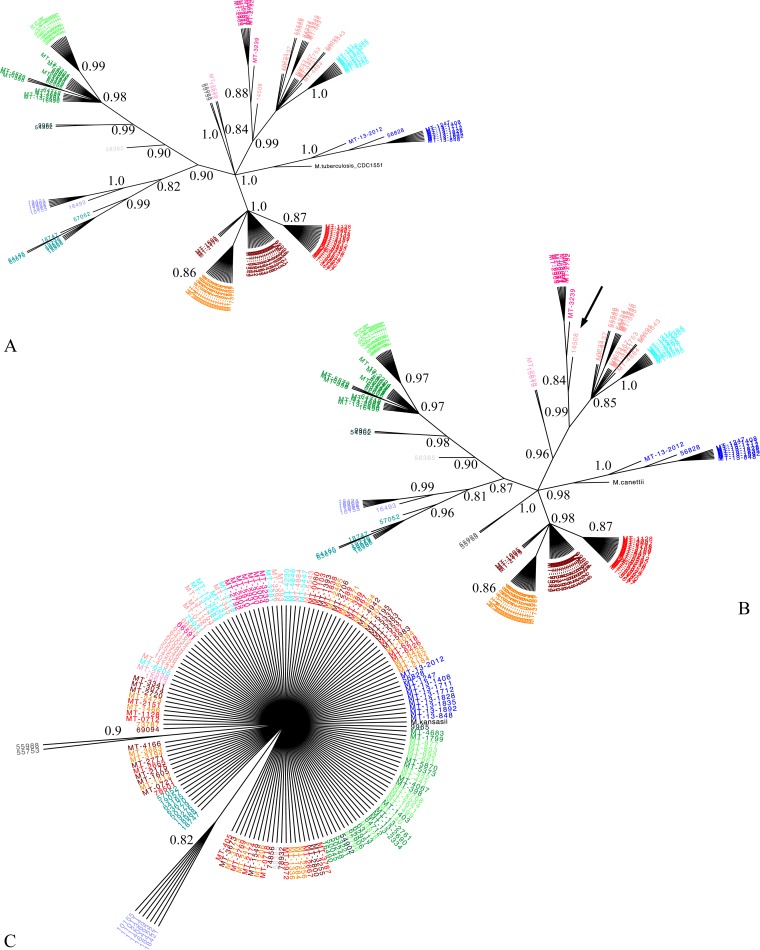
FIG 1.
Impact of reference genome choice on phylogeny. Maximum likelihood trees with 1,000 bootstrap replicates. Branches of <80% bootstrap threshold are collapsed (branch lengths are therefore not to scale). For clarity, bootstrap P values are indicated up to the most proximal node defining each cluster. Isolates were colored for their respective clusters, identified according to CDC1551 (and H37Rv) (8). Isolates were then kept the same color across all panels to facilitate quick comparison between the new reference analysis and CDC1551 (see Table S4 in the supplemental material for cluster names). (A) Reference M. tuberculosis lineage 4 CDC1551, using the Tamura 3 parameter model of nucleotide substitution with 1,522 SNP loci (19). (B) Reference M. canettii, using the general time-reversible (GTR) model of nucleotide substitution with 17,406 SNP loci (20). Using M. canettii as a reference, a single isolate changed clusters (arrow). (C) Reference M. kansasii, using the GTR model of nucleotide substitution with 34,127 SNP loci.