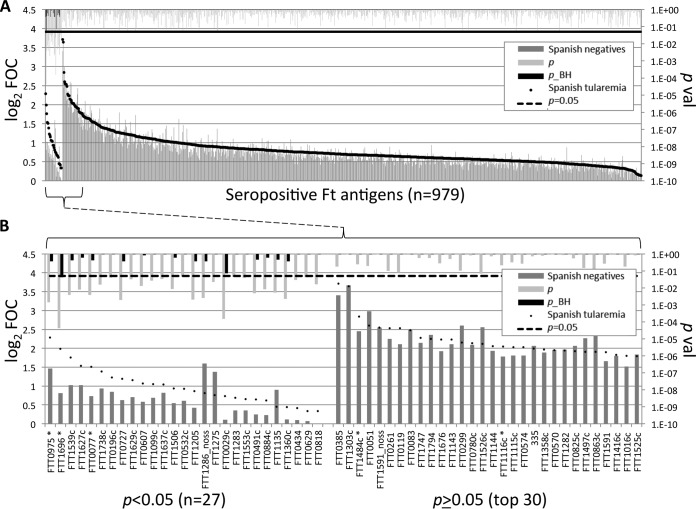
FIG 3.
Identification of antigens associated with F. tularensis exposure using proteome microarrays. The microarrays were probed with sera from Spanish tularemia cases (sera obtained from groups 1 to 3 at the second time point) and Spanish individuals negative for tularemia (sera obtained from groups 4 and 5 at the second time point), and IgG was visualized by indirect immunofluorescence (see Materials and Methods). A serum sample was defined to be positive for an antigen if the array signal was >2 times the median for the IVTT reaction control spots in at least 10% of the samples from either group. (A) Antigens to which the serum samples were reactive (n = 979). Ft, F. tularensis. (B) Enlargement of the 27 discriminatory antigens and top 30 nondiscriminatory antigens from panel A. Array data were normalized by dividing the IVTT reaction protein spot intensity by the sample-specific median for the 77 IVTT reaction control spots printed throughout the chip to give the FOC and then taking the base 2 logarithm of the ratio (log2 FOC). *, antigens identified to be significant in the work of Sundaresh et al. (25); noss, the signal sequence was deleted; p_BH, P values corrected for false discovery using the Benjamini-Hochberg method (31).