Figure 1. miRNA profiling of stomach metaplastic lineages.
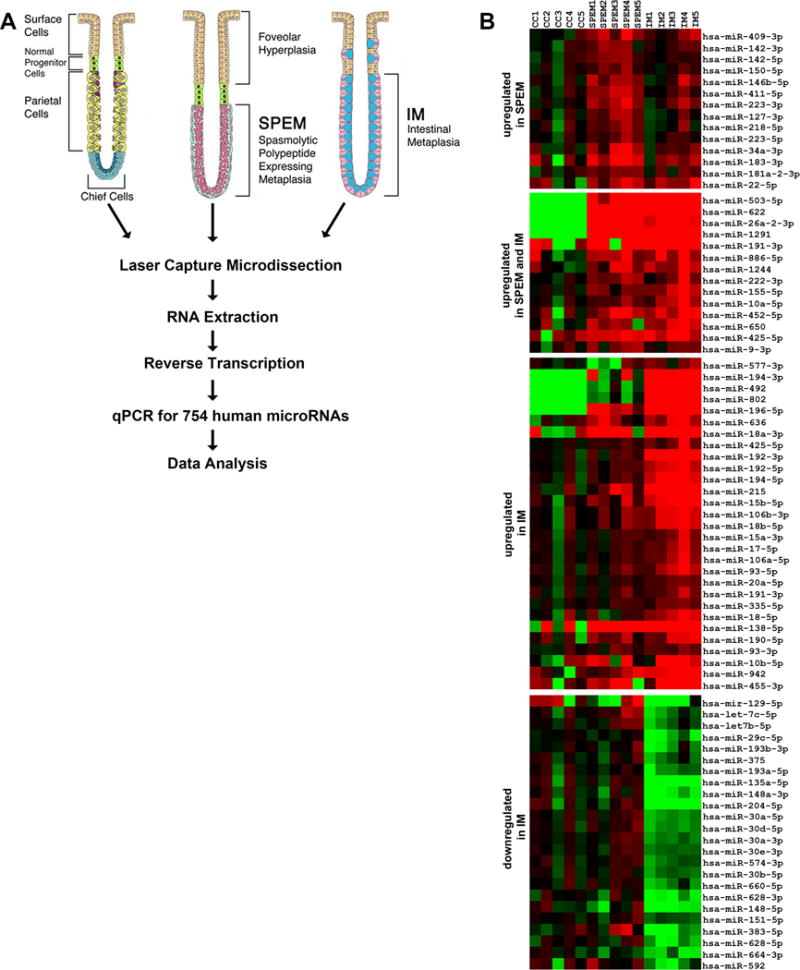
(A) Method schematics. SPEM and intestinal metaplasia lesions (five cases each) were microdissected from 10 μm sections of frozen biopsies from gastric resections of intestinal type tumors in the fundus. As a control comparison group, chief cells were microdissected from normal stomach mucosa. Total RNA was extracted using the mirVana kit (Applied Biosystems) and then, 60 ng of RNA from each sample were converted to cDNA and pre-amplified using Megapool RT and pre-amplification primers (Applied Biosystems). MicroRNA expression was determined by Real Time PCR using TaqMan MicroRNA Arrays (Applied Biosystems). (B) Venn diagram summarizing the overlap of miRNA changes, both up-regulated and down-regulated, in SPEM and intestinal metaplasia (IM). (C) Heat map of the expression profiles of microRNAs differentially expressed in the two types of metaplasias in the human stomach (intestinal metaplasia and SPEM) in comparison with normal chief cells.
