Abstract
Microfluidics, featuring microfabricated structures, is a technology for manipulating fluids at the micrometer scale. The small dimension and flexibility of microfluidic systems are ideal for mimicking molecular and cellular microenvironment, and show great potential in translational research and development. Here, the recent progress of microfluidics in biological and biomedical applications, including molecular analysis, cellular analysis, and chip-based material delivery and biomimetic design is presented. The potential future developments in the translational microfluidics field are also discussed.
1. Introduction
Microfluidics, featuring microfabricated structures, is a technology for manipulating fluids at the micrometer scale. It is developed from semiconductor industry and usually referred to as micro-total-analytical (microTAS) system or lab-on-a-chip (LOC) device.[1] The core elements of microfluidics are channels, chambers, and valves, which can be integrated to perform precise and complicated operations. Compared to conventional analytical tools, microfluidics offer a number of unique merits, including reduced sample consumptions, high throughput, high speed analysis, and improved sensitivity.[2] Nowadays, it has shown the potential to produce powerful and robust biomedical devices.
Microfluidics is characterized by the unique feature of small dimensions, which make it easy to transfer mass and heat in microchannels, and thus enable faster reaction and detection. Small dimensions also lead to reduced consumption of samples and reagents and greatly decreases analysis cost. The reduced dimensions also make it possible to fabricate small devices, such as point-of-care (POC) devices, to achieve on-site testing. Microfluidic systems can also integrate multiple processes such as purification, separation, and detection to miniaturize complex laboratory procedures onto a small device and to achieve multiplex analysis with high throughput and high sensitivity. Due to the unique features and advantages of microfluidic systems, microfluidics has impacted a range of applications, including chemistry, engineering, biology, medicine, and other fields.[3]
In this review, we will not include the exciting applications in other fields such as chemistry and engineering but focus on the biological and clinical applications of the microfluidic systems. The small dimension and flexibility of microfluidic systems are eligible to control molecular and cellular microenvironment and show great potential in translational research and development. We highlight only the recent 5-year progress in molecular analysis (protein and nucleic acids markers), cellular analysis (cell mechanics, cell migration, cell separation and sorting, and single cell analysis), and material delivery and biomimetic design (chip-based material delivery and organ-on-chip for drug discovery). The outputs of these studies and their clinical implementations demonstrate that microfluidics is an enabling technology for biological analysis and shows great translational potential in clinical analysis and therapeutics. We will also discuss the translational challenges encountered in current research. Finally, this is not a comprehensive review. Although we tried hard to include all relevant articles, some interesting and important research may still be missing due to limited space and our limited expertise in the field.
2. Molecular Analysis
In tissues and biological fluids, there are numerous proteins, functioning as enzymes, hormones, signals, and materials.[4] Nucleic acids, including DNA and RNA, function as a storage medium of genetic information, which is necessary to synthesize proteins.[5] In clinical diagnosis, the levels of proteins and nucleic acids in biological fluids, such as blood, can be used as biological markers of certain diseases.[6] Detection and analysis of these biomarkers are defined as molecular diagnostics and many new platforms have been developed and used to diagnose and monitor disease, assess risk, and decide therapy for individual patients. Traditional technologies for the detection of biomarkers rely on sophisticated equipment, highly trained operators, large sample size, and are generally time-consuming and expensive, and thus are limited to laboratory use. It is critical to develop small and portable devices that enable rapid, accurate, and sensitive analysis. The portable devices may be useful for on-site detection. Microfluidic platforms have the potential to overcome some of these limitations. The major advantages of microfluidics in molecular diagnostics are that they can achieve more sensitive and faster detection due to the improved integration, improved automation, reduced contamination, reduced sample size, and reduced consumed reagents.
2.1. Protein Detection
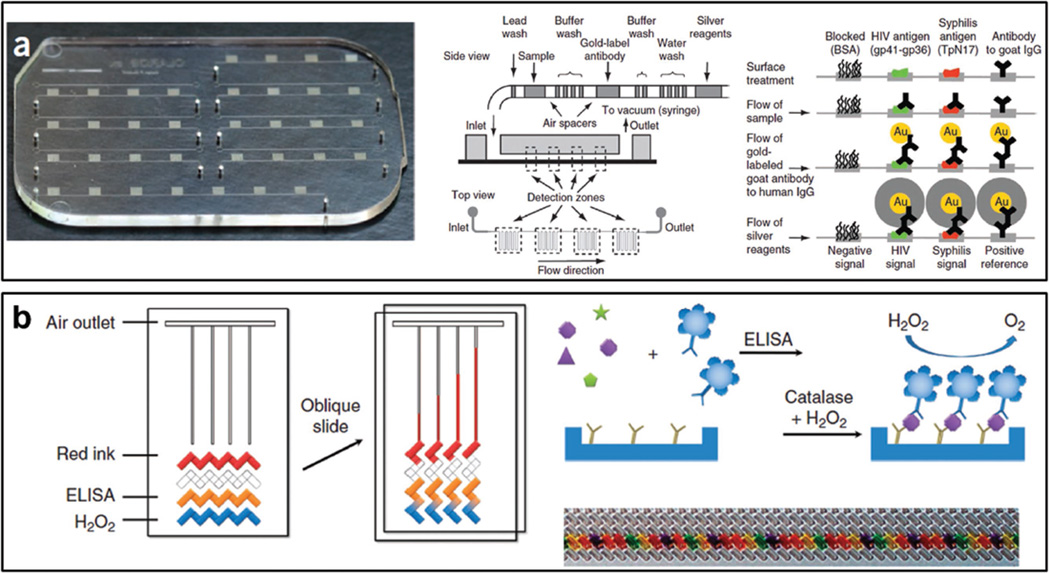
Enzyme-linked immunosorbent assay (ELISA) is currently the “gold standard” for protein detection. It uses antibody and color change to identify specific protein markers, and has been widely used as a diagnostic tool in clinical laboratories.[7] The traditional ELISA requires several steps of washing and incubation with reagents. A recent study demonstrated by Chin et al.[8] shows that it is possible to miniaturize these steps into a small microfluidic chip (Figure 1a). The microfluidic chip (mChip) was loaded with a sequence of wash buffer and reagents over antibody coated detection zone. The mChip was demonstrated to detect human immunodeficiency virus (HIV) antigen in 1 µL of unprocessed whole blood, and the reaction can be completed within 20 min. Compared to the benchtop ELISA, the mChip requires minimal equipment. To further integrate the functions of readout, data processing and data communication, the same research group next used a smartphone to achieve these.[9] The microfluidic ELISA assay was coupled into a dongle that is attached to a smartphone to perform triplexed detection of HIV and syphilis. The combination of hardware, software, and microfluidic specifications in a smartphone generates a more friendly user interface. Though these devices show potential for POC diagnosis, they have limitations such as low throughput, semiquantitative detection.
Figure 1.
Microfluidic devices for protein detection. a) Photo of microfluidic chip and schematic illustration of loading of multiple reagents to detect HIV and syphilis antigens. Reproduced with permission.[8] Copyright 2011, NPG. b) Schematic illustration of volumetric bar-chart chip. Protein concentrations are converted into travel distance of ink bars by ELISA reaction. Reproduced with permission.[11c] Copyright 2012, NPG.
Diseases often lead to the changes of multiprotein levels in biospecimens. Thus multiprotein detection will provide better diagnostic results. Microfluidic devices feature fluid control in multichannels, and have shown powerful applications in multiplex protein detection. The reported integrated blood barcode chip (IBBC) provides a good approach for multiplex protein analysis.[10] The IBBC chip, designed with multiple DNA-encoded antibody barcode arrays, can achieve the plasma separation from whole blood and the following rapid measurement of a panel of protein markers over a broad concentration range. Recently, another example for multiplexed protein detection is volumetric bar-chart chip (V-Chip),[11] which is first developed by our group. The V-Chip converts protein concentrations into travel distance of ink bar by ELISA reactions (Figure 1b)[11c] and allows visual quantification of multiplex protein markers. The sensitivity of V-Chip can be improved by using nanoparticles.[12] The capability of V-Chip for multiplex and quantitative analysis of protein biomarkers makes it a powerful POC platform in resource limited settings. However, the sensitivity and analysis speed of V-Chip can be further improved. The mass production of V-Chip should be also considered in the future.
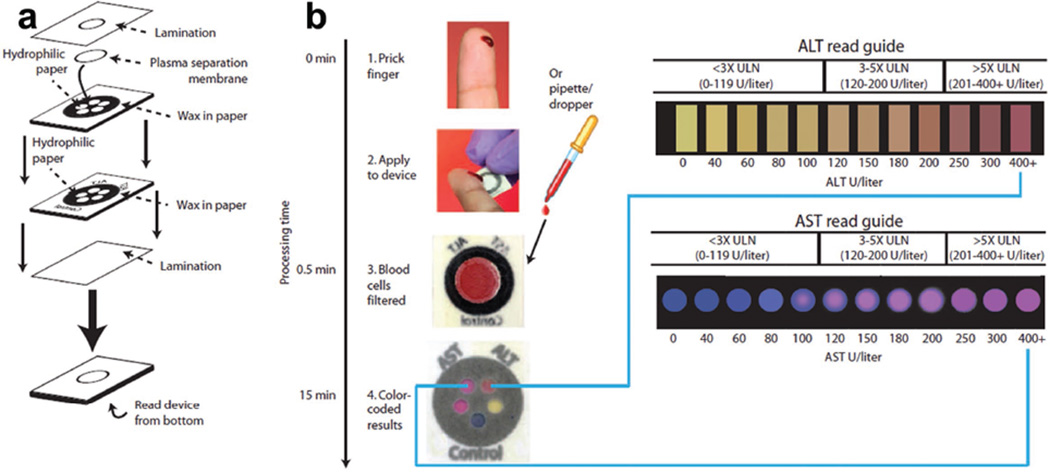
Paper, or a flexible cellulose-based material, has recently been used to fabricate microfluidic chips, which have the advantages of lightweight, ease-of-use, biocompatibility, and low cost. These properties enable paper-based microfluidic chips appropriate for molecular analysis in on-site detection and POC applications. ELISA assay has been demonstrated in paper-based microfluidic chips with the combined advantages of sensitivity and specificity of ELISA, and the low cost and ease-of-use of paper chips. Paper-based microfluidic chips have shown powerful applications to detect a number of biomarkers, including HIV antigen, influenza virus antigen and tumor markers.[13] Recently, Pollock et al. developed a paper-based microfluidic device, consisting of multilayered structures, to achieve plasma separation and colorimetric measurement of liver disease markers aspartate aminotransferase and alanine aminotransferase in a semiquantitative way (Figure 2).[13b] One possible disadvantage of paper-based microfluidics is that the reaction, such as ELISA, is less sensitive than conventional ELISA, which may possibly affect the detection limit. For a more detailed description of paper microfluidics for molecular detection, we refer the reader to the specialized review published by Xia et al.[14]
Figure 2.
Paper-based microfluidic device for ALT and AST assays. a) The device consists of multilayer papers and membranes. b) AST and ALT test zones are matched to color read guide to obtain protein concentrations. Reproduced with permission.[13b] Copyright 2012, AAAS.
2.2. Nucleic Acids Detection
The detection of nucleic acids to obtain sequence-specific genetic information has broad applications ranging from fundamental research to disease diagnosis. To date, it is still challenging to achieve rapid and accurate detection of nucleic acids in biological samples. Microfluidics technology, capable of integrating multiple biochemical processes in a single device, offers a promising tool for such applications.
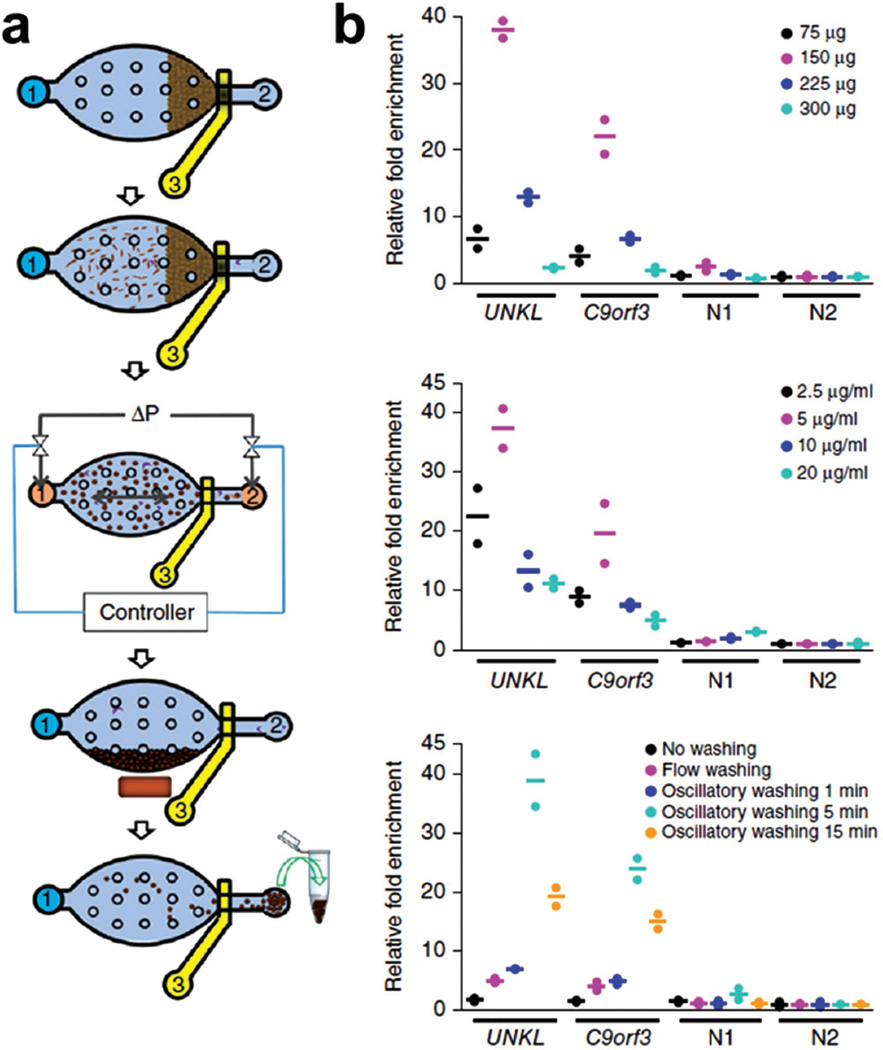
To minimize the adverse effect of complex background in biological samples, the isolation and purification of nucleic acids is a crucial step and holds the key for most genetic analysis.[15] Microfluidic technology has been combined with bead technology for the isolation of nucleic acids.[15b] A recent study by Cao et al.[15a] shows that the combination of the two technologies can achieve high-efficiency collection of DNA for epigenomic analysis (Figure 3). This study introduces a microfluidic oscillatory washing-based ChIP-seq to purify DNA in as few as 100 cells, compared to 107 cells needed in conventional ChIP-seq technique. Using this technique, some new super enhancers were found in hematopoietic stem and progenitor cells, and this indicates the powerful potential application in epigenomic profiling. However, the use of beads for nucleic acids isolation usually has the problem of instability.
Figure 3.
Microfluidic device for the collection of DNA in as few as 100 cells. a) Schematic illustration of microfluidic oscillatory washing-based ChIP-seq to purify DNA. The five major steps are formation of a packed bed of beads, flowing of chromatin fragments, oscillatory washing, removal of the unbound chromatin, and collection of beads. b) Optimization of protocol including the amount of beads, concentration of antibody used for coating and washing duration. Reproduced with permission.[15a] Copyright 2015, NPG.
The detection of nucleic acids is usually achieved using biomolecular techniques such as polymerase chain reaction (PCR) and DNA hybridization,[16] which can be integrated with sample preparation and target purification in a microfluidic chip. Ferguson et al.[16a] developed a magnetic integrated microfluidic electrochemical detector, which can perform immunomagnetic target capture, concentration, purification, PCR amplification, and electrochemical detection of influenza H1N1 from throat swab samples. Our group recently reported a multistage propelled volumetric bar chart chip (MV-Chip) to detect DNA.[16e] DNA hybridization in MV-Chip introduced catalase initiator to start propellant reaction. After three-stage cascade amplification on platinum-deposited films, the detection sensitivity can be improved by 1000 fold. The results can be read out by ink bar charts with naked eyes. Digital PCR is a new approach for nucleic acids detection, and can also be performed in a single microfluidic chip with the advantage of increasing detection sensitivity. Shen et al.[16c] has used slip chip technology-based digital PCR to detect Staphylococcus aureus genomic DNA with the throughput of 1280 detection units. This technique provides a simple strategy to count nucleic acids. Though these methods show improved sensitivity and feasibility, the high cost still limits them as laboratory prototypes.
3. Cellular Analysis
3.1. Biophysical Analysis
Quite a few human diseases, such as arthritis, asthma, cancer, malaria, and sickle cell anemia, are not only related to the molecular alterations, but also associated with abnormalities in the structural and physical characteristics of cells.[17] Studying mechanical properties of cells associated with these diseases and their correlation with molecular changes could provide better understanding of the disease occurring and progression.[18] Physical properties of cells include cell morphology (cell and nuclear shape, nuclear architecture, cell surface), cell mechanics (deformability, stiffness, and adhesion), and cell migration.[19] Here we focus on the study of cell mechanics and cell migration as well as how microfluidic techniques can contribute to these applications.
3.1.1. Cell Mechanics
Cell mechanical properties, including deformability, stiffness, and adhesion, can be used as label-free biomarkers to indicate cytoskeletal and nuclear changes in various diseases. For example, it has been reported that cancer cells are more flexible than normal cells and that decreased cell stiffness is correlated with increased metastatic potential.[20] Compared to molecular markers, label-free mechanical markers may have the potential to reduce analysis cost and time in diagnosis and disease treatment.
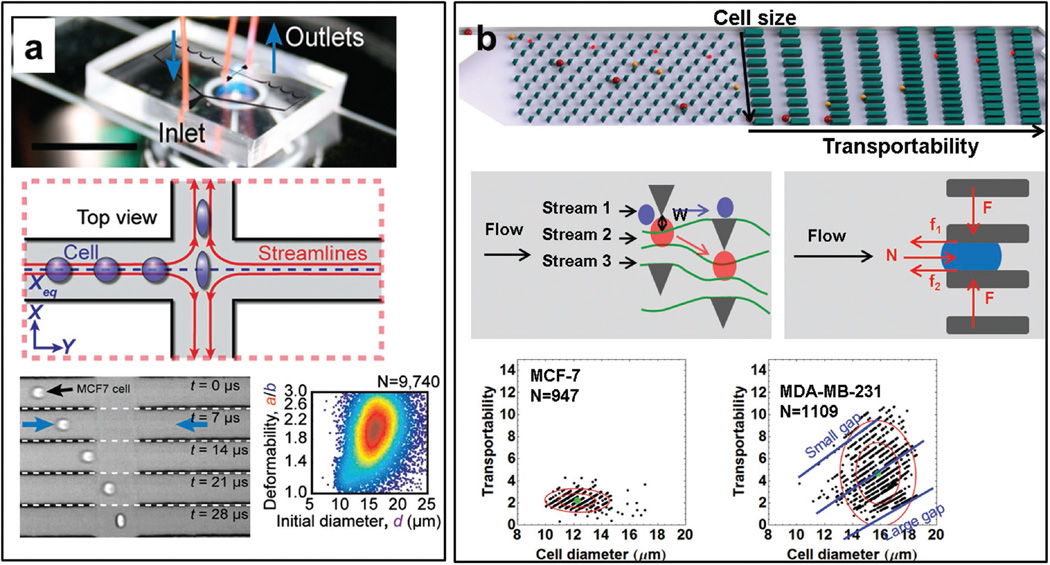
Traditionally, a variety of techniques, such as atomic force microscopy (AFM), micropipette aspiration, magnetic tweezers, and optical stretching, have been used to probe cell mechanical properties.[17,20a] However, these techniques are usually time-consuming and labor intensive. To overcome these disadvantages and improve the analysis throughput, a lot of microfluidic systems have been developed. Gossett et al.[21] introduced a deformability cytometry (DC) chip to characterize single cell mechanical property (Figure 4a). Single cells were stretched by inertial force and then analyzed by a high-speed camera. A cytometry-like map was next obtained by plotting cell diameter and deformability. The DC chip was used to distinguish populations of cells within blood and pleural fluids in a total number of several thousand.[21,22] To further improve the throughput, Guck et al. developed a real-time deformability cytometry (RT-DC),[23] which achieved measurement of more than 100 000 cells in one experiment. The RT-DC platform was used to study the effect of cell cycles on deformability and differentiate blood cells and their precursors. In our group, we developed a microfluidic cytometric (MC) platform to separate cells by size and the ability to pass through small gaps (Figure 4b).[24] We found that decreased cell stiffness and cell-surface frictional forces may help to cancer cells passing through narrow spaces in metastasis, which was also reported in the study of Byun et al.[25]
Figure 4.
Microfluidic mechanical analysis of cells. a) Schematic illustration of deformability cytometry. A high-speed camera is used to take images of stretched cells in microchannels. Density scatter plot of size and deformability is then got. Reproduced with permission.[21] Copyright 2012, National Academy of Sciences. b) Microfluidic cytometric analysis of cancer cell transportability. The device is composed of deterministic lateral displacement and trapping barrier structures. Cancer cells are separated by their size and transportability through small gaps. Transportability and diameter of each cell is plotted. Reproduced with permission.[24] Copyright 2015, NPG.
These studies indicate that microfluidic techniques have powerful potential for the measurement of cell mechanical properties. However, cells in vivo are frequently subjected to various mechanical stimuli in the physiological environment. The devices should be designed to mimic these stimuli in a more controllable way. Moreover, it is still difficult using current methods to carry out downstream molecular analysis following characterization of cell mechanics. Our group have used a microfluidic separation chip to isolate flexible cells from a SUM 149 cell line, and found tumor-initiating cells are enriched in flexible cells.[26] Such studies are particularly important to explore the correlation between biophysical markers and molecular markers.
3.1.2. Cell Migration
Cell migration plays an important role in various physiological processes, such as embryogenesis, tissue repair, cancer metastasis, and inflammation. For example, in cancer metastasis, cancer cells gain invasiveness at the primary site, migrate through extracellular matrix, circulate in blood or lymphatic system, and interact with the microenvironment at the secondary site.[27] So cancer cell migration is critical for metastasis. Inflammation is also characterized by the migration of leucocytes into tissue.[28] Migration of cells is usually regulated by chemical or physical cues. Microfluidics can be used to produce microchamber or micropillar structures, which mimic the physiological cues of cell migration in a more controlled way.
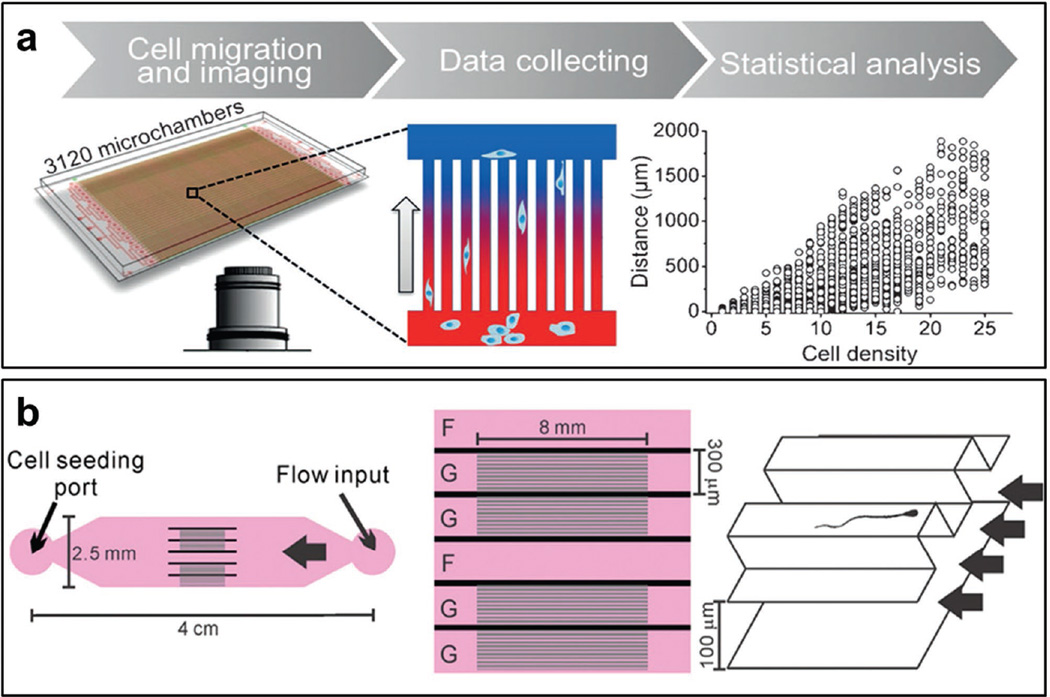
Chemotaxis is the phenomena that migration of cells is induced by the gradient of extracellular chemicals, such as chemokines and growth factors. In our group, we have developed a microfluidic chip to study the chemotaxis behavior of cancer cells.[29] The microfluidic chip is composed of 3120 microchambers, and used to screen antimetastatic drugs for their inhibition of mesenchymal migration and prevention of metastatic malignancy (Figure 5a). Our group further developed a high-throughput microdevice with 4000 microwells to monitor real-time, 3D cancer cell migration, and allow the chip to screen drugs that inhibit cell migration.[30] Chemotaxis induced leucocyte migration plays an important role in inflammation. Boneschansker et al. recently introduces a microfluidic device using two microarrays of microchannels to simultaneously analyze four qualitative migration patterns: chemo-attraction, -repulsion, -kinesis, and -inhibition.[31] This device provides an avenue for precise characterization of leucocyte migration.
Figure 5.
Microfluidic devices for cell migration. a) Schematic illustration of a microfluidic chip for high throughput analysis of cancer cell migration for drug screening. Chemogradient is generated in each microchamber. Reproduced with permission.[29] Copyright 2014, Wiley-VCH. b) Schematic illustration of a microfluidic chip to mimic the biophysical environment of female reproductive tract and to study the migration behavior of sperms and pathogens. Reproduced with permission.[34] Copyright 2015, National Academy of Sciences.
Cell migration is not only regulated by chemicals, but also affected by biophysical cues, such as fluid flow. In solid tumors, there is interstitial flow (IF) from tumor core. Recently, a microfluidic chip is developed to mimic this phenomena and study the effect of fluid pressure on cell migration in collagen I type hydrogels.[32] The results show that IF induces the activation of β1 integrin adhesion complexes localizing into the upstream side of the cell. The same group also developed microfluidic-based assays to study cancer cell intravasation,[33] which is a critical step in cancer metastasis. The microfluidic chip can reproduce the tumor-vascular interface in three dimensions, allowing for precise simulation of endothelial barrier function. In fertilization, sperm cells migrate against a gentle flow and pass through uterotubal junction to reach the egg. The surface microgrooves and fluid flows critically regulate sperm cell migration. A microfluidic chip was developed to mimic the biophysical environment of female reproductive tract and to study the migration behavior of sperms and pathogens in a paper published by Tung et al (Figure 5b).[34] The results indicate that the fluid flow and microgrooves facilitate sperm migration, but prevent pathogen migration.
These studies demonstrate that microfluidics is a powerful tool to study cell migration by mimicking the biochemical and biophysical cues. However, current studies are mainly focused on the characterization. It is challenging to collect large population of cells based on the differences of migration potential and perform downstream molecular analysis.[35] Besides microchambers, micropillar structures have also been used to study mechanotransduction and cell migration. Micropillars can not only support cell adhesion and spreading but also act as force sensors to map live-cell contractile forces. Recent studies have reported the use of micropillar structures to direct cell differentiation, proliferation, and migration.[36] Reffay et al.[36c] used the traction forces exerted by micropillars to study collective cell migration behavior. The results show that leader cells exert a large traction force mediated by RhoA signaling.
3.2. Cell Separation and Sorting
Isolation and sorting of cells from complex mixtures is often used to enrich or purify well-defined cell populations in biology, medicine, and clinical applications. For example, patients with cancers or other disorders of blood and immune systems can be treated by sorting and transplanting hematopoietic stem cells.[37] To date, the most widely used cell sorter is fluorescence-activated cell sorting (FACS),[38] which can achieve multiplexed analysis and a throughput of 50 000 cells per second. Microfluidic devices, which are capable of precise spatial and temporal control of fluid flow and cell manipulation, have been recently used for cell isolation and sorting,[39] such as circulating tumor cells, stem cells, and lymphocytes.
Circulating tumor cells (CTCs) are tumor cells that disseminate from primary cancers and spread through circulation system. Studies have indicated that CTC levels in blood are indication of tumor progression and survival, and can be regarded as a noninvasive liquid biopsy.[40] The detection of CTCs will provide valuable information for both basic and clinical research. However, CTCs are extremely rare, at the level of 1–100 CTCs in over 1 × 109 blood cells, which makes the detection of CTCs challenging. The current gold standard of CTCs detection is CellSearch, which uses magnetic particles functionalized with antiepithelial cell adhesion molecule (EpCAM) for positive capture.[41] To improve the capture efficiency, EpCAM antibodies were modified onto microchannel surface in microfluidic devices.[42] To further improve the capture efficiency and throughput, some physical structures, including chaotic[43] and geometrically enhanced differential immunocapture structures,[44] were used.
One possible disadvantage of affinity-based CTCs capture is that CTCs may have undergone epithelial-mesenchymal transition (EMT) and have downregulated expression of epithelial markers.[45] And it is also difficult to recover the captured CTCs. Regarding that CTCs have bigger cell diameter and are stiffer than leukocytes, many microfluidic devices have been developed to capture CTCs based on the physical differences between CTCs and blood cells. These microfluidic devices utilized filtration,[46] hydrodynamic force,[47] and inertial focusing[48] for CTCs isolation, which often achieve high capture yield and high throughput. Certain features inside microfluidics channel can significantly enhance the CTC capture efficiency. A notable development of double spiral microchannel has contributed a significant high CTC capture efficiency and made label free detection possible.[47d] In the study of Liu et al., deterministic lateral displacement (DLD) structure was used for cancer cell enrichment with the throughput up to 9.6 mL min−1.[47b] Because the size of CTCs (15–25 µm) are overlapped with leukocytes (5–20 µm), it is difficult to improve the capture purity in physical separation techniques.
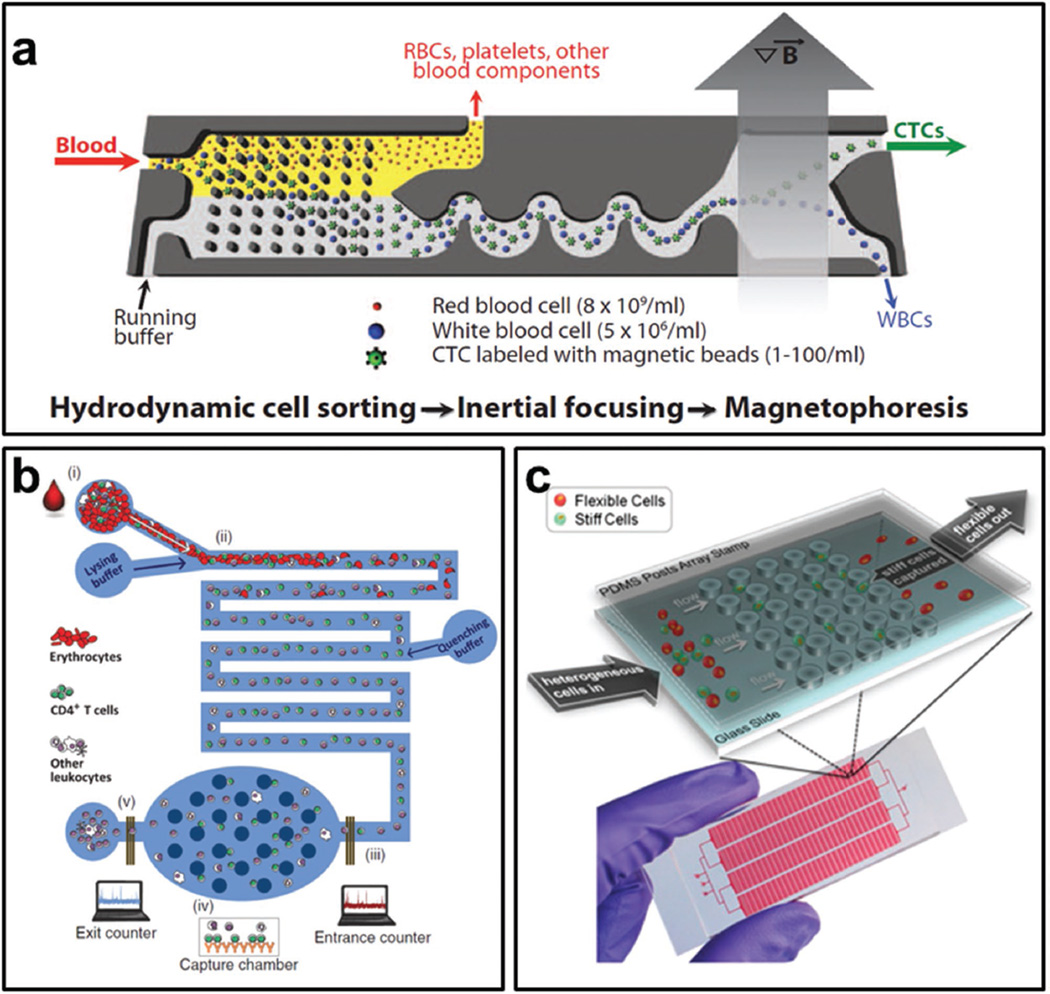
More recently, a lot of studies have been reported to use integrated microfluidic devices for CTCs capture. In Toner’s recent study, they developed a microfluidic chip, composed of DLD, inertial focusing and magnetic separation structures to capture CTCs (Figure 6a).[48] Leukocytes were depleted in this device and the throughput was up to 107 cells per second. The same group also reported that CTCs clusters captured in a microfluidic chip have increased metastatic potential.[49] These studies successfully demonstrate microfluidics is a promising technique for CTCs isolation and sorting. Further studies should focus on the downstream analysis of CTCs, such as gene expression, protein expression and drug resistance, which may be then correlated with patient outcomes and demonstrate the usefulness of CTCs detection in clinical. For a more comprehensive review of microfluidics for CTCs isolation, we refer the reader to the reviews published by Green et al.[50] and Hyun et al.[51]
Figure 6.
Cell separation and sorting. a) Schematic illustration of a microfluidic chip utilizing hydrodynamic sorting, inertial focusing, and magnetic separation to capture CTCs. Reproduced with permission.[48] Copyright 2013, AAAS. b) A microfluidic device is coated with antibodies for capture and enumeration of HIV-related CD4+ and CD8+ leukocytes. Reproduced with permission.[52] Copyright 2013, AAAS. c) Schematic illustration of a microfluidic chip to sort flexible cells using trapping barrier structure. Cancer stem cells are enriched in flexible cell population. Reproduced with permission.[26] Copyright 2012, National Academy of Sciences.
Besides CTCs, microfluidic devices have also been used for other cells’ isolation and sorting such as leukocytes and stem cells for biological research or clinical diagnostics. In HIV disease, the absolute count of CD4+ and CD8+ leukocytes is routinely used for diagnostic management. Microfluidic devices can be modified with these antibodies for the capture and enumeration of HIV-related CD4+ and CD8+ leukocytes (Figure 6b).[52] Such approaches have the potential to be developed as simple HIV diagnostic tools. Stem cells are undifferentiated cells with the potential to differentiate into specialized cells, and can be used to treat or prevent a disease or condition, which is called stem cell therapy and requires high purity stem cells. Although FACS can purify undifferentiated populations using specific surface markers, the survival rate of sorted cells is low due to single cell dissociation. Singh et al. introduced a microfluidic chip to sort human pluripotent stem cells by exploiting the adhesion strength differences of undifferentiated and differentiated stem cells.[53] The human pluripotent stem cells were enriched to 95%–99% purity with over 80% survival. Cancer cells are highly heterogeneous. Our group developed a microfluidic cell separation chip to isolate flexible cells from a cancer cell line by squeezing cells through micropost arrays (Figure 6c).[26] It was found cancer stem cells were enriched in the flexible populations. These examples indicate that microfluidic sorting based on cellular biochemical and biophysical signatures will provide novel biological techniques as well as new diagnostic tools. However, the throughput, purity and viability of sorted cells should be further improved.
3.3. Single Cell Analysis
Cells are heterogeneous even within small cell populations. Individual cells differ in size, expressed RNA and protein levels, the variations of which play an important role in many biological processes, including cancer progression, embryonic development, immune responses, and neuron networking.[54] Single cell analysis such as genome sequencing, RNA sequencing, and protein expression, is required to better understand these biological processes. Single cell analysis is also required for scarce samples with limited numbers of cells, e.g., CTCs, embryo samples and primary neurons.
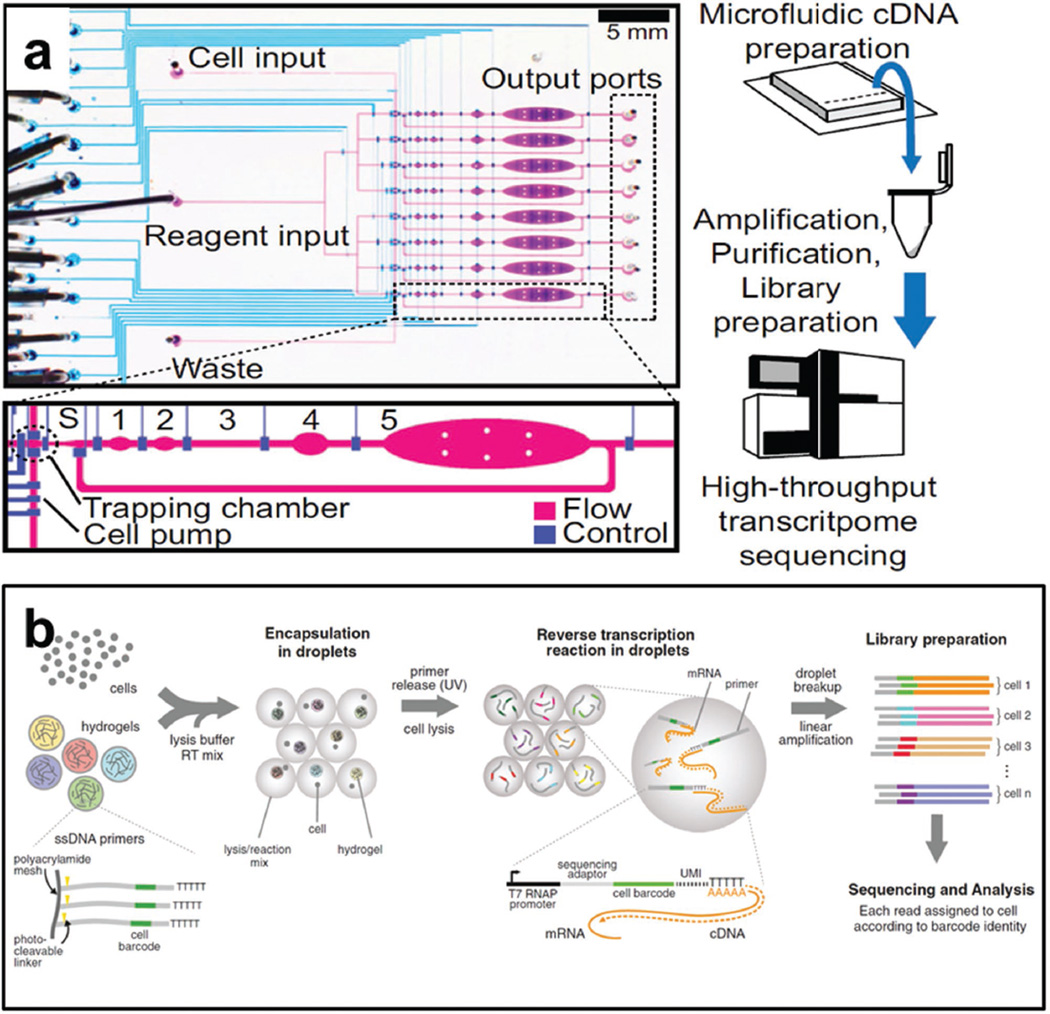
To perform single cell analysis, the first and key step is the isolation of single cells. The current single cell isolation approaches are serial dilution, micromanipulation, fluorescence-activated sorting, and laser-capture microdissection.[55] Recently, microfluidic chips have been used for single cell isolation. Our group developed a hand-held single-cell pipet (hSCP) that can achieve rapid, efficient, and unbiased isolation of single cells (Figure 7).[56] A hook is designed in the hSCP to capture and release single cells. The isolated single cells can be easily transferred into standard 96-/384-well plates for downstream biochemical assays. Microfluidic chips are not only capable of single cell isolation, but also have the ability to perform molecular analysis, such as genomics, transcriptomics, and protein expression assay. Because vast majority of genome sequence is considered to be relatively stable, it is mainly introduced here how microfluidic technology contributes to the heterogeneity study of transcriptomics and protein expressions. Transcriptomic analysis is usually performed by first amplification of cDNA followed by high-throughput transcriptome sequencing (RNA-Seq).[57] The single cell cDNA preparation and amplification can be achieved in microfluidic chips. The Fluidigm has developed a C1 platform for such application. In this platform, microchannels and microvalves are used to construct individual reaction chambers. After single cells are isolated, cells are automatically lysed and template is quickly prepared for qPCR or sequencing analysis. Wu et al. reported that the cDNA preparation and amplification in C1 platform show improved detection sensitivity than conventional tube-based approaches.[58] Recently, Steets et al. used similar microfluidic chambers to perform reverse transcription and cDNA amplification (single-cell RNA-Seq technology, developed by Tang et al.[59]) of embryonic stem cells with minimized contamination and improved detection sensitivity (Figure 8a).[60] Besides microchambers, droplets have also been used to capture single cells and perform RNA sequencing. Klein et al. (Figure 8b)[61] and Macosko et al.[62] separately developed droplet microfluidic platforms to encapsulate single cells with DNA encoded microhydrogels and microbeads. The encoded droplets make it possible to analyze RNA transcripts from thousands of individual cells while remembering transcripts’ cell of origin, which can significantly decrease detection cost. With this technique, in the analysis of mouse embryonic stem cells, single-cell heterogeneity revealed population structure and gene regulatory linkages, while in the study of retina, 39 retinal cell populations were identified.
Figure 7.
Hand-held single-cell pipet (hSCP) for single cell isolation. a) Schematic illustration of hSCP. The hSCP tip has a hook to capture single cell. b) Demonstration of single cell isolation. c,d) Tip dimensions and work flow for single-cell isolation. Reproduced with permission.[56] Copyright 2014, American Chemical Society.
Figure 8.
Single-cell transcriptomic analysis. a) Schematic illustration of chip structure and experimental protocol. Fluid flow is controlled by microvalves. cDNA is collected from chip, and further amplified for sequencing. Reproduced with permission.[60] Copyright 2014, National Academy of Sciences. b) A droplet microfluidic platform for sequencing and analysis of thousands of single cells. cDNA in each droplet is tagged with a barcode for sequencing. Reproduced with permission.[61] Copyright 2015, Elsevier.
Microchambers, microwells, and microdroplets have also been used to study single cell protein expression or secreted proteins. Protein expression assays perform multiplex protein measurements and connect genomic information to biological functions.[63] To achieve multiplex protein detection, some approaches, e.g., single-cell barcode chips (SCBCs), have been developed. The SCBCs features miniature antibody arrays, which can be encoded with DNA oligomer. Barcoded antibody array in microchambers or microwells have been used to detect secreted, cytoplasmic, and membrane proteins.[64] The small volume (nL) of solution in microenvironment can enrich proteins to allow standard immunoassay detection. Ma et al. reported the use of SCBCs to profile intracellular signaling pathways in single tumor cells and measure secreted proteins from phenotypically single T cells, which may have potential application in immune monitoring and clinical assessment (Figure 9a).[64c] Recently, Lu et al. introduced a barcode platform with spatial and spectral encoding antibodies for detection of 42 immune effector proteins secreted from single immune cells, representing the highest multiplexing recorded to date for a single-cell secretion assay.[64b] Droplet microfluidic chips have also been used to encapsulate single cells into droplets for protein assay. Wang et al. developed a droplet microfluidic chip to phenotype single cells based on the products they secrete or consume.[65] Individual cells were compartmentalized for growth and analysis in nanoliter aqueous droplets to identify scarce xylose-overconsuming Saccharomyces cerevisiae cells. Ng et al. developed a digital microfluidic platform for cell culture, stimulation, and immunocytochemistry in single droplets, which can achieve high time resolution for screening signaling responses of a heterogeneous cell population (Figure 9b).[66] Protein assay sorting in droplets has also been achieved. Mazutis et al. presented a droplet microfluidics chips to compartmentalize single cells in droplets enabling high throughput analysis and sorting of single cells based on the proteins released from or secreted by single cells.[67]
Figure 9.
Single-cell protein analysis. a) Single-cell barcode chips (SCBCs) for multiplex protein detection. Concentration of secreted proteins from individual cell is measured in microchamber. Reproduced with permission.[64c] Copyright 2011, NPG. b) A digital microfluidic platform for cell culture, stimulation, and immunocytochemistry in single droplets. This platform can achieve high time resolution for screening signaling responses of a heterogeneous cell population. Reproduced with permission.[66] Copyright 2015, NPG.
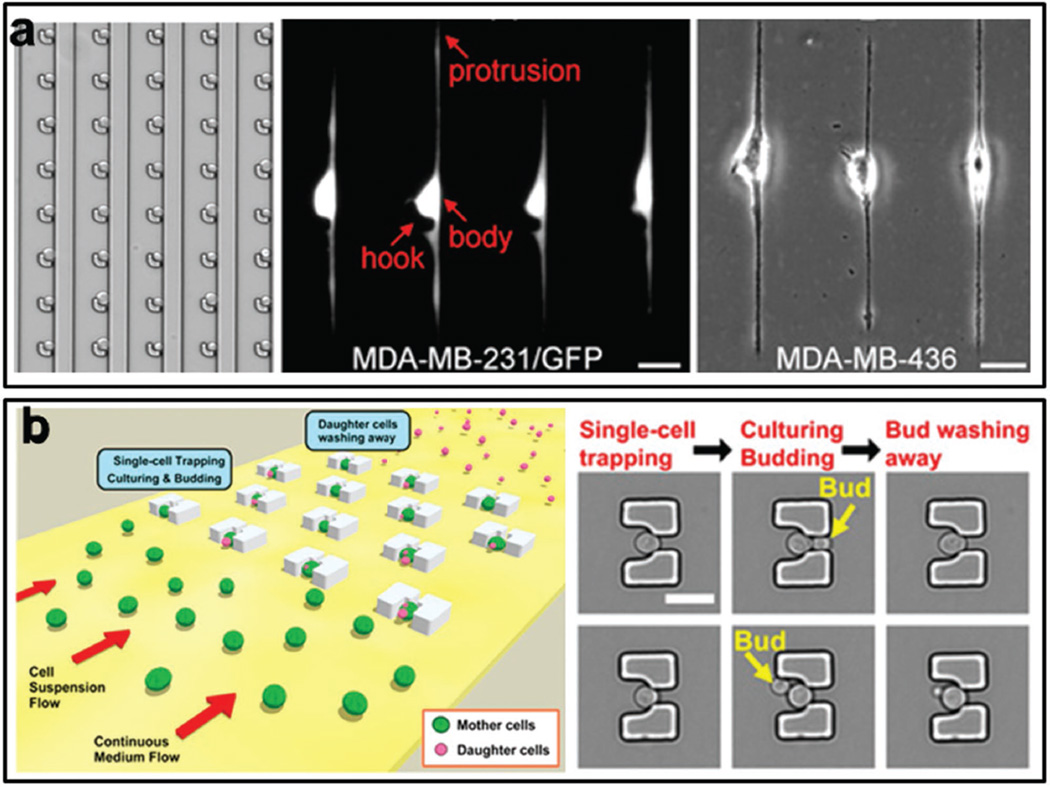
Microfluidic chips can also be used to study morphological phenotypes of single cells, such as protrusion and cell aging. Our group has developed a unique live-cell printing technique, termed “Block-Cell-Printing” (BloC-Printing), which employs hooks to capture and hold cells at designated positions for functional assay of single cell biological behaviors (Figure 10a).[68] With this technique, we studied single cell protrusion, which characterizes cells’ capability to extend the membrane and plays an important role in cell migration and invasion. The results show that breast basal-like cancer cells had greater membrane elongation ability than breast luminal-like cancer cells. Recently, our group also developed a microfluidic platform to study aging of yeast cells.[69] Aging and age-associated diseases have shown global impacts on public health. In the previous aging study of yeast cells, it is difficult to track the entire lifespan of mother cells, which are mixed with the daughter cells. In our developed platform, the single mother cells were trapped in microstructures and it was easy to observe the budding behavior of each mother cell because the daughter cells can be washed away (Figure 10b). Thus our approach provides an efficient avenue for aging study of yeast cells.
Figure 10.
Morphological analysis of single cells. a) Block-Cell-Printing (BloC-Printing) for single cell protrusion analysis. A hook is used to capture single cell and the length of cell protrusion is measured. Reproduced with permission.[68] Copyright 2014, National Academy of Sciences. b) Trapping of single yeast cell for aging analysis. Single mother cells are trapped in microstructures and it is easy to observe the budding behavior of each mother cell because the daughter cells can be easily washed away. Reproduced with permission.[69] Copyright 2015, National Academy of Sciences.
Due to the unique advantages of fluid handling and cell manipulation, microfluidics is particularly suitable for studying single cell-cell communications. Traditionally, cell–cell communication is studied in petri dish with random cell distribution. Skelley et al. developed a microfluidic device to pair cells and study their fusion behavior.[70] The pairing efficiency was up to 70%. The same group next used similar microfluidic design to pair lymphocytes and study their activation dynamics (Figure 11a).[71] Recently, our group introduced a vertical cell pairing system to study the dynamic synapses between cancer cells and NK cells (Figure 11b).[72] The immunological synapse communication is critically important in immune cells. Compared to the horizontal approaches, the vertical pairing allows high-resolution imaging.
Figure 11.
On-chip cell–cell communication. a) Schematic illustration and images of lymphocytes pairing and activation dynamics in microfluidic channel. Reproduced with permission.[71] Copyright 2015, NPG. b) Vertical pairing of cancer cell and NK cells. The immunological synapse communication is studied. Compared to the horizontal approaches, the vertical pairing allows high-resolution imaging.[72] Copyright 2015, The American Association of Immunologists.
These studies indicate that microfluidics has provided an efficient avenue for single cell heterogeneity analysis. However, the current microfluidic single-cell analysis is mainly limited for fundamental research due to the low throughput and high analysis cost. Therefore, further improvements including the increase of throughput, efficiency, and accuracy and the decrease of analysis cost, are required for translational applications.
4. Material Delivery and Biomimetic Design
4.1. Chip-Based Drug Testing and Material Delivery
Microfluidic technologies have advanced the drug delivery filed, including drug carrier synthesis and screening platforms from cellular to organism level.[73] One such application is the integration of a concentration gradient generator with a cell culture platform, which makes drug testing more efficient and accurate. The most common designs are tree-like gradient generators (TLGGs).[74] To enable high-throughput drug screening, combinatorial gradient generators have also been developed. However, drug screening on cellular level is not so efficient to discover drugs for treating patients. To better mimic in vivo drug effects, tissue-on-a-chip devices are developed and can reflect more physiologically in vivo behavior of the drug. Moreover, combined with micro particles and nanoparticle as drug carries, the drug delivery has achieved targeting and releasing specifically and effectively in the tissue and organism levels. Recently, more and more attentions have been paid on organ-on-a-chip devices, which are more suitable models for drug delivery and screening.[75] In addition, implanted microfluidic drug delivery devices have also been developed for many disease models.[76]
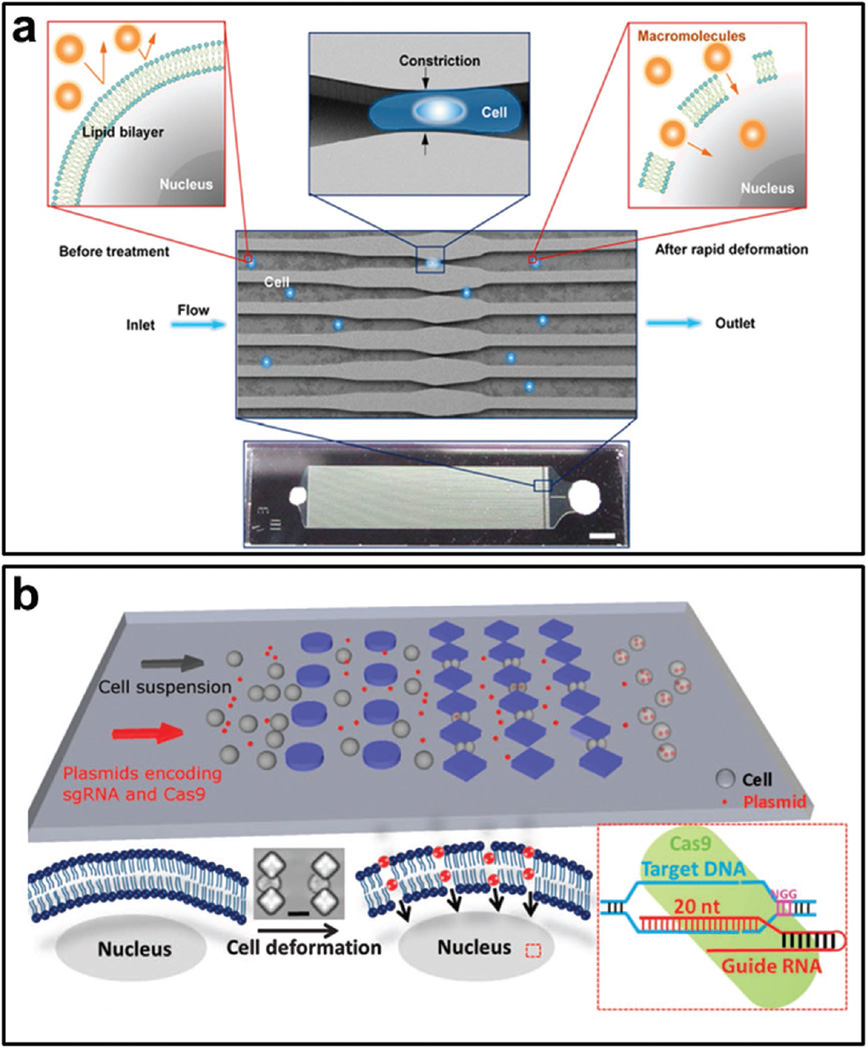
Recently, a new mechanical delivery method has been developed utilizing microfluidic technology.[77] Through rapid mechanical deformation of cells to produce transient membrane holes, the delivery materials can diffuse into cell cytosol. This method has achieved delivery of almost any macromolecule of interest to almost any cell type in a high throughput manner. Sharei et al. reported the delivery of a range of materials such as carbon nanotubes, proteins, and siRNA (Figure 12a).[77b] They also achieved the delivery of transcription factors for cell reprogramming. With the optimization of the microfluidic chip, our group has achieved the delivery of new types of nucleic acids such as ssDNA, siRNAs, and large-sized plasmids, which will enhance the applicability of this technology in both research and therapeutics (Figure 12b).[77a] Moreover, by combining CRISPR/Cas9 technologies with the unique membrane deformation delivery method, we have achieved genome editing and gene disruption, which may provide an efficient platform for applying gene editing in biomedical research and gene-targeting therapy.
Figure 12.
Chip-based material delivery. a) Schematic illustration of cell deformation in microchannels for material delivery. When cells pass through microchannels, the rapid deformation generates membrane holes enabling the delivery from outside to inside. Reproduced with permission.[77b] Copyright 2013, National Academy of Sciences. b) The same mechanism enables the delivery of CRISPR-Cas9 into cells for gene editing. Reproduced with permission.[77a] Copyright 2015, AAAS.
This novel microfluidics based delivery method has the great potential in cell therapy including therapeutic genome editing for blood disorders and cancer immunotherapy. One biggest advantage of this mechanical delivery method is broad applicability across different cell types, particularly hard-to-transfect cells. One important direction is to achieve efficient and nontoxic delivery of Cas9 and sgRNAs to human hematopoietic stem cells. By combining the CRISPR-Cas9 knockin system and the microfluidic delivery chip to correct the disease gene mutation, it will be possible to correct disease mutation genes by gene targeting therapy. Another application is cancer T-cell therapy. Generation of tumor-targeted human T lymphocytes endowed with therapeutic properties is an important strategy toward cancer therapy.[78] CAR (chimeric antigen receptors) T-cell therapy has experienced three generations to make the CAR-modified T cells more effective and therapeutic toward cancer. However, a high level of virus-free CAR genes delivery into T cells is still a big challenge. Hence this microfluidics based delivery system provides a new strategy for delivering CARs into T cells with high efficiency.
4.2. Biomimetic Design
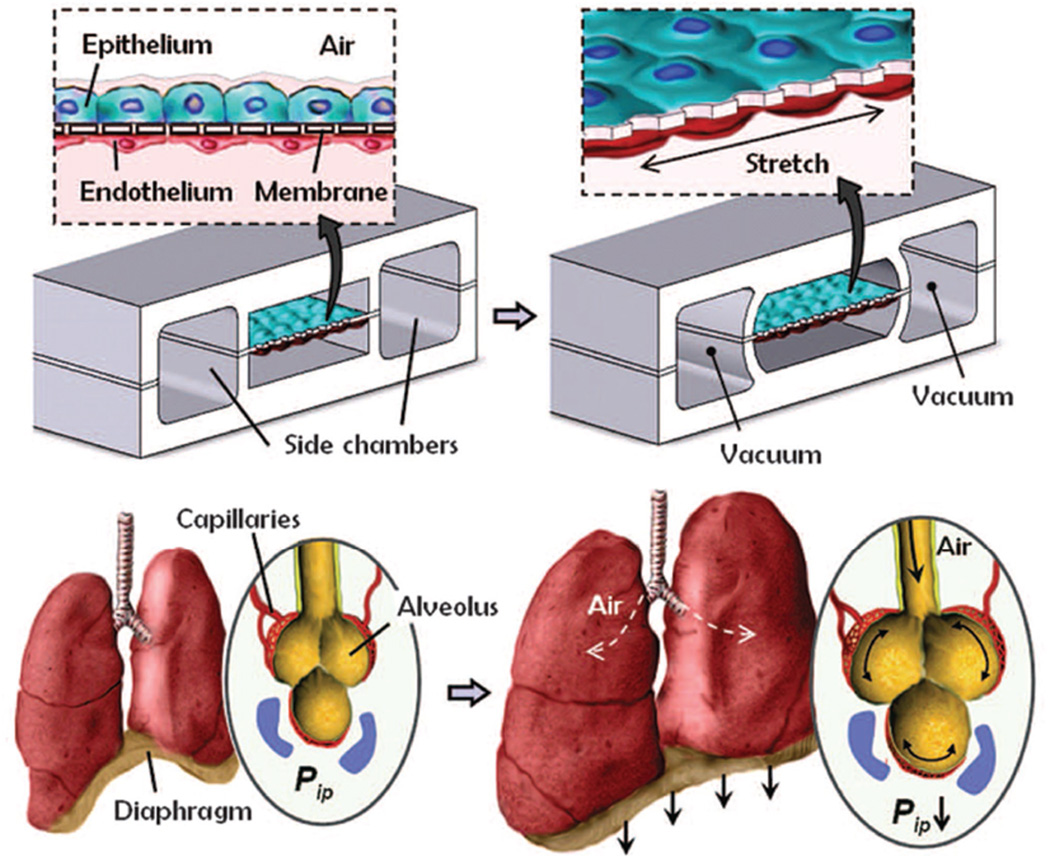
Organs-on-chips are the most powerful biomimetic designs in microfluidics field. Microfluidic devices are designed to culture living cells in continuously perfused, micrometer sized chambers to mimic physiological functions of tissues and organs. By recapitulating tissue and organ functions, organs-on-chips have been widely used in disease modeling and drug screening. With the great potential in the investigation of organ physiology and disease, this technology has been used to mimic functions of liver, kidney, intestine, lung, heart, fat, bone, skin, cornea, nerve, marrow, blood vessels, and so on.[75,79] Moreover, combined with conventional 3D culture systems such as hydrogel-based methods and bioreactors, organs-on-chips have proved very useful for studying tissue- and organ-level behaviors. Huh has developed a reconstituting organ-level human lung model on a chip and reproduced complex integrated organ-level responses to bacteria and inflammatory cytokines introduced into the alveolar space (Figure 13).[79d] Later on, they have developed a human disease model of drug toxicity induced pulmonary edema in a lung-on-a-chip device.[79c] Another bone marrow-on-a-chip model has also been developed by the same group to replicate hematopoietic niche physiology in vitro.[79g] The engineered bone marrow (eBM) retains hematopoietic stem and progenitor cells in normal in vivo-like proportions for at least 1 week in culture.
Figure 13.
Lung-on-a-chip microdevice to mimic lung function. Compartmentalized PDMS microchannels enable breathing-like movements of membranes by applying vaccum. Reproduced with permission.[79d] Copyright 2010, AAAS.
Organs-on-chips can be used for drug screening and development. Shuler group made the first microfluidic cell-culture analog of a mathematical pharmacokinetics (PK) model for drug development and toxicology.[80] The purpose of the microfluidic model is to study adsorption, distribution, metabolism, elimination, and toxicity (ADMET) of chemicals entirely in vitro rather than in animals. Other studies have also explored the organs-on-chips models to study drug development like livers-on-chips for drug metabolism. Moreover, the microfluidic organs-on-chips models are also suitable for cancer drug mentalism, drug transport and toxicity analysis. Sophisticated human organs-on-chips may help to discover new biomarkers of drug efficacy, toxicity, or disease response, which could benefit clinical trials.
Biomaterial fabrication for tissue regeneration is another application in biomimetic design.[81] By combining injecting ability and micro porosity, Griffin et al. have provided a unique biomaterial scaffold for efficient cellular network formation in vitro and bulk tissue integration in vivo, which would enable novel routes for tissue regeneration field.[81a]
In recent years, the development of induced pluripotent stem (iPS) technology[82] has given microfluidic organs-on-chips researches unlimited potential. The iPS technology can produce more kinds of cells which can be used in generation of organs-on-chips models. What is more, CRISPR-Cas9 system based gene editing technologies have been proved a strong method to introduce specific diseases mutations and generate diseases models, and if it is combined with organs-on-chips platforms, it could be very useful to develop drug screening toward diseases particular in human models. We also refer the readers to recent comprehensive reviews of organs-on-chips published by Polini et al.,[83] Esch et al.,[84] and Bhatia et al.[79b]
5. Challenges and Perspectives
The unique features and advantages of microfluidics, such as small dimensions, faster reaction, and reduced analysis cost, have led to a plethora of powerful and robust devices, which impact biological and biomedical fields. Up to now, there are thousands of published microfluidics papers every year. Though significant progress has been achieved, there are still a lot of challenges facing the microfluidic field. Here, we will briefly discuss the three challenges: microfluidics materials, integration, and in vivo-oriented simulation. Addressing these challenges will promote the translational applications of microfluidic systems. Regarding these challenges, we also refer the readers to the reviews published by Nge et al.[2] and Sackmann et al.[1b]
5.1. Microfluidics Materials
Microfluidic devices can be prepared from different materials, including silicon, glass, PDMS, paper, and membrane. The first-generation microfluidics materials are silicon and glass, which can be processed by standard machining or etching.[85] This process is usually time-consuming and labor-intensive and requires expensive facilities. Though these microfluidics materials can be used for some applications such as capillary electrophoresis, slip-chip, and digital PCR, the lack of mass-production and high fabrication cost limit their commercialization and translational applications.
Up to now, the most common microfluidics material is PDMS, which is optically transparent, gas- and water-permeable. PDMS molds are elastomers fabricated by soft lithography and can be reversibly or irreversibly bonded to glass, PDMS, and other materials.[86] The optically transparent property enables ease of observation and imaging while the gas- and water-permeable property can create suitable conditions for cell culture. These properties together with the ease of fabrication and reasonable cost have led to a plethora of microfluidic designs. Though PDMS devices have been extensively used for academic research, there are only few commercialized products due to the lack of scalability. It is also reported that PDMS devices are found to leach uncrosslinked agents, which are harmful in the translational applications.[87]
To overcome the limitations of PDMS and facilitate the commercialization, microfluidics community has explored to use thermoplastics for device fabrication. Thermoplastics, e.g., polystyrene[88] and cyclic olefin copolymer,[8] are cheap materials with excellent light transmission property. These plastics can be processed by laminate, embossing, and injection molding, which make mass production possible. In translational applications, thermoplastics like polystyrene may be more acceptable than PDMS by biologists due to the long history as cell culture materials. Besides thermoplastics, cheap materials such as paper, wax, and membranes have also been used as microfluidics materials for POC applications in resource-limited settings.
The selection of appropriate materials can not only reduce the analysis cost, but also helps to improve stability and accuracy, which is another challenge for clinical use of microfluidic devices. PDMS is a kind of widely used microfluidic materials. However, the hydrophobicity results in easy protein adsorption and thus decreases detection limit and stability. To overcome these disadvantages, it is often required to do surface modification or use other materials, such as glass.
5.2. Integration
Though a variety of microfluidics devices were developed for the clinic or biological research, only few of them have been widely used or commercialized. The major challenges include the integration and automation of devices. Integration refers to the combination of different components, e.g., pumps, valves, mixers, and detectors into a single device to complete multiple processes such as sample pretreatment, reaction, and detection. Since each component varies a lot in the design and function, it is extremely challenging to not only connect them together but also make them compatible with each other. Integration provides the advantages of miniaturization, automation, ease of use, and low-cost assay, which are critical for translational applications.
Here, we introduce several excellent examples of microfluidics integration. In CTCs isolation, preliminary enrichment is often required to improve the throughput due to the extreme rarity of CTCs in blood cells. The enrichment and the following separation of CTCs can be integrated into one chip. Ozkumur et al.[48] and Karabacak et al.[89] reported a microfluidic chip with three functional structures for CTCs isolation. The first structure consisted of DLD arrays and was used to enrich large diameter cells that include CTCs and leukocytes. The second structure was inertial focusing and used to focus the enriched cells to the center of channel. In the third structure, there was a magnetic field, which was used to separate magnetic beads-labeled leukocytes. The three-stage negative isolation technique achieved high throughput, high recovery and high purity isolation of CTCs. Deformability cytometry presents another example of integrated devices for automated analysis of cell mechanics. Traditionally, cell mechanics can be studied using a number of analytic methods, such as AFM, micropipette aspiration, magnetic tweezer, and optical stretching. These methods are usually time-consuming and labor-cost. Gossett et al.[21] and Otto et al.[23] separately reported microfluidics-based deformability cytometers, which can not only measure cell deformability but also integrate data acquisition for automated analysis. Similar to fluorescence-based flow cytometer, the output results are easy to understand and interpret. The demonstrated capability for diagnosing malignant pleural effusions and phenotyping blood cells suggests the potential application in clinic.
The introduction of multiple steps can also improve stability and accuracy of the analysis. For example, sample preparation usually involves separating and concentrating of targets. The manual preparation of these processes depends on the skill level of each person, and possibly decreases the stability, while the integration of these operations can easily achieve automation and improves the stability. To further improve the accuracy and stability, the integration of other technologies, such as nanotechnology, can be considered. V-Chip was first developed by our group for protein or nucleic acid detection. To increase the detection limit and accuracy, nanoparticles[12b] or nanoporous substrates[11d] have been used. In the test of biomarkers of nonsmall cell lung cancer, the detection limit can be tenfold improved over the original V-Chip.[11d] The high surface-to-volume ratios of nanomaterials can efficiently increase the number of binding sites for antibodies and enable the low detection limit.
Besides integration, simple operation should be also considered to develop robust devices. Sometimes, the simplest may be the best. Our group has developed a handed single-cell pipette[56] to isolate single cells. The device is composed of a microchannel and a hook and can be connected to a pipette tip. This simple and user-friendly approach may be more accepted by biologists or clinicians.
5.3. In-Vivo-Oriented Simulation
Many microfluidics devices have been developed to study the biological behaviors of cells, tissues, and organs. These microfluidics models are required to more closely mimic the in vivo environment or functions. Organ-on-chips provide excellent examples of using microfluidics devices to recapitulate tissue and organ functions for disease modeling and drug screening. Though a lot of in vitro models such as lung-on-a-chip and bone marrow-on-a-chip have been developed, they can only recapsulate some of the physiological architectures due to the complexity of human organs. Future work should focus on how to improve in vivo-oriented simulation by optimizing microfluidic design, selecting biomimetic materials and integrating more components.
Acknowledgments
The authors are grateful for funding from NIH-R01CA180083, NIH-U54CA143837, NIH-R01DA035868, NIH-R56AG049714, NIH/NCI (1R21CA191179-01A1) and Golfers against Cancer Foundation.
Biographies

Zongbin Liu is a postdoctoral fellow in Department of Nanomedicine at Houston Methodist Research Institute. He received his Ph.D. degree in Biomedical Engineering from The Hong Kong Polytechnic University in 2010. Dr. Liu’s current research interests are: circulating tumor cells’ isolation and analysis, microfluidic mechanical analysis of cancer cells, and PEG-based biomaterials for tissue engineering and drug delivery.

Xin Han is a postdoctoral fellow in Department of Nanomedicine at Houston Methodist Research Institute. He received his Ph.D. degree in Biochemistry and Molecular Biology from Sun Yat-Sen University in 2013. Dr. Han’s research focuses on telomere and aging, cancer metastasis, and translational medicine in microfluidics.

Lidong Qin is a professor in Department of Nanomedicine at Houston Methodist Research Institute. Dr. Qin received his Ph.D. in Chemistry from Northwestern University, Evanston, Illinois and completed a postdoctoral traineeship in Cancer Nanotechnology at the California Institute of Technology. Dr. Qin’s research focus is microfluidics biotechnology.
Contributor Information
Dr. Zongbin Liu, Department of Nanomedicine, Houston Methodist Research Institute, Houston, TX 77030, USA Department of Cell and Developmental Biology, Weill Medical College of Cornell University, New York, NY 10065, USA.
Dr. Xin Han, Department of Nanomedicine, Houston Methodist Research Institute, Houston, TX 77030, USA Department of Cell and Developmental Biology, Weill Medical College of Cornell University, New York, NY 10065, USA.
Prof. Lidong Qin, Email: lqin@houstonmethodist.org, Department of Nanomedicine, Houston Methodist Research Institute, Houston, TX 77030, USA; Department of Cell and Developmental Biology, Weill Medical College of Cornell University, New York, NY 10065, USA; Department of Molecular and Cellular Oncology, The University of Texas M. D. Anderson Cancer Center, Houston, TX 77030, USA.
References
- 1.a) Kovarik ML, Ornoff DM, Melvin AT, Dobes NC, Wang Y, Dickinson AJ, Gach PC, Shah PK, Allbritton NL. Anal. Chem. 2013;85:451. doi: 10.1021/ac3031543. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sackmann EK, Fulton AL, Beebe DJ. Nature. 2014;507:181. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 2.Nge PN, Rogers CI, Woolley AT. Chem. Rev. 2013;113:2550. doi: 10.1021/cr300337x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitesides GM. Nature. 2006;442:368. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 4.Anderson NL, Anderson NG. Mol. Cell. Proteom. 2002;1:845. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 5.Levy RM. Science. 1988;241:234. doi: 10.1126/science.241.4862.234. [DOI] [PubMed] [Google Scholar]
- 6.Song YJ, Huang YY, Liu XW, Zhang XJ, Ferrari M, Qin LD. Trends Biotechnol. 2014;32:132. doi: 10.1016/j.tibtech.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hornbeck PV. Curr. Protoc. Immunol. 2015;110:2.1.1. doi: 10.1002/0471142735.im0201s110. [DOI] [PubMed] [Google Scholar]
- 8.Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umviligihozo G, Karita E, Mwambarangwe L, Braunstein SL, van de Wijgert J, Sahabo R, Justman JE, El-Sadr W, Sia SK. Nat. Med. 2011;17:1015. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 9.Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, Cadinu P, Meng F, Chee NH, Kim J, Chin CD, Munyazesa E, Mugwaneza P, Rai AJ, Mugisha V, Castro AR, Steinmiller D, Linder V, Justman JE, Nsanzimana S, Sia SK. Sci. Transl. Med. 2015;7:273re271. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 10.Fan R, Vermesh O, Srivastava A, Yen BK, Qin L, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Nat. Biotechnol. 2008;26:1373. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Li Y, Xuan J, Song Y, Wang P, Qin L. Lab Chip. 2015;15:3300. doi: 10.1039/c5lc00529a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Li Y, Xuan J, Xia T, Han X, Song Y, Cao Z, Jiang X, Guo Y, Wang P, Qin L. Anal. Chem. 2015;87:3771. doi: 10.1021/ac504301y. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Song Y, Zhang Y, Bernard PE, Reuben JM, Ueno NT, Arlinghaus RB, Zu Y, Qin L. Nat. Commun. 2012;3:1283. doi: 10.1038/ncomms2292. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Li Y, Xuan J, Song Y, Qi W, He B, Wang P, Qin L. ACS Nano. 2016;10:1283. doi: 10.1021/acsnano.5b07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Song Y, Xia X, Wu X, Wang P, Qin L. Angew. Chem. Int. Ed. 2014;53:12451. doi: 10.1002/anie.201404349. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhu Z, Guan Z, Jia S, Lei Z, Lin S, Zhang H, Ma Y, Tian ZQ, Yang CJ. Angew. Chem. Int. Ed. 2014;53:12503. doi: 10.1002/anie.201405995. [DOI] [PubMed] [Google Scholar]
- 13.a) Lei KF, Huang CH, Kuo RL, Chang CK, Chen KF, Tsao KC, Tsang NM. Anal. Chem. Acta. 2015;883:37. doi: 10.1016/j.aca.2015.02.071. [DOI] [PubMed] [Google Scholar]; b) Pollock NR, Rolland JP, Kumar S, Beattie PD, Jain S, Noubary F, Wong VL, Pohlmann RA, Ryan US, Whitesides GM. Sci. Transl. Med. 2012;4:152ra129. doi: 10.1126/scitranslmed.3003981. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Warren AD, Kwong GA, Wood DK, Lin KY, Bhatia SN. Proc. Natl. Acad. Sci. USA. 2014;111:3671. doi: 10.1073/pnas.1314651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia Y, Si J, Li Z. Biosens. Bioelectron. 2016;77:774. doi: 10.1016/j.bios.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 15.a) Cao ZN, Chen CY, He B, Tan K, Lu C. Nat. Methods. 2015;12:959. doi: 10.1038/nmeth.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Reinholt SJ, Baeumner AJ. Angew. Chem. Int. Ed. 2014;53:13988. doi: 10.1002/anie.201309580. [DOI] [PubMed] [Google Scholar]; c) Root BE, Agarwal AK, Kelso DM, Barron AE. Anal. Chem. 2011;83:982. doi: 10.1021/ac102736g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Ferguson BS, Buchsbaum SF, Wu TT, Hsieh K, Xiao Y, Sun R, Soh HT. J. Am. Chem. Soc. 2011;133:9129. doi: 10.1021/ja203981w. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kim TH, Park J, Kim CJ, Cho YK. Anal. Chem. 2014;86:3841. doi: 10.1021/ac403971h. [DOI] [PubMed] [Google Scholar]; c) Shen F, Du WB, Kreutz JE, Fok A, Ismagilov RF. Lab Chip. 2010;10:2666. doi: 10.1039/c004521g. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Shen F, Sun B, Kreutz JE, Davydova EK, Du WB, Reddy PL, Joseph LJ, Ismagilov RF. J. Am. Chem. Soc. 2011;133:17705. doi: 10.1021/ja2060116. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Song YJ, Wang YC, Qin LD. J. Am. Chem. Soc. 2013;135:16785. doi: 10.1021/ja4085397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Carlo D. J. Lab. Autom. 2012;17:32. doi: 10.1177/2211068211431630. [DOI] [PubMed] [Google Scholar]
- 18.Lee GY, Lim CT. Trends Biotechnol. 2007;25:111. doi: 10.1016/j.tibtech.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Physical Sciences–Oncology Centers Network. Agus DB, Alexander JF, Arap W, Ashili S, Aslan JE, Austin RH, Backman V, Bethel KJ, Bonneau R, Chen WC, Chen-Tanyolac C, Choi NC, Curley SA, Dallas M, Damania D, Davies PC, Decuzzi P, Dickinson L, Estevez-Salmeron L, Estrella V, Ferrari M, Fischbach C, Foo J, Fraley SI, Frantz C, Fuhrmann A, Gascard P, Gatenby RA, Geng Y, Gerecht S, Gillies RJ, Godin B, Grady WM, Greenfield A, Hemphill C, Hempstead BL, Hielscher A, Hillis WD, Holland EC, Ibrahim-Hashim A, Jacks T, Johnson RH, Joo A, Katz JE, Kelbauskas L, Kesselman C, King MR, Konstantopoulos K, Kraning-Rush CM, Kuhn P, Kung K, Kwee B, Lakins JN, Lambert G, Liao D, Licht JD, Liphardt JT, Liu L, Lloyd MC, Lyubimova A, Mallick P, Marko J, McCarty OJ, Meldrum DR, Michor F, Mumenthaler SM, Nandakumar V, O’Halloran TV, Oh S, Pasqualini R, Paszek MJ, Philips KG, Poultney CS, Rana K, Reinhart-King CA, Ros R, Semenza GL, Senechal P, Shuler ML, Srinivasan S, Staunton JR, Stypula Y, Subramanian H, Tlsty TD, Tormoen GW, Tseng Y, van Oudenaarden A, Verbridge SS, Wan JC, Weaver VM, Widom J, Will C, Wirtz D, Wojtkowiak J, Wu PH. Sci. Rep. 2013;3:1449. doi: 10.1038/srep01449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a) Cross SE, Jin YS, Rao J, Gimzewski JK. Nat. Nanotechnol. 2007;2:780. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]; b) Cross SE, Jin YS, Tondre J, Wong R, Rao J, Gimzewski JK. Nanotechnology. 2008;19:384003. doi: 10.1088/0957-4484/19/38/384003. [DOI] [PubMed] [Google Scholar]
- 21.Gossett DR, Tse HT, Lee SA, Ying Y, Lindgren AG, Yang OO, Rao J, Clark AT, Di Carlo D. Proc. Natl. Acad. Sci. USA. 2012;109:7630. doi: 10.1073/pnas.1200107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tse HT, Gossett DR, Moon YS, Masaeli M, Sohsman M, Ying Y, Mislick K, Adams RP, Rao J, Di Carlo D. Sci. Transl. Med. 2013;5:212ra163. doi: 10.1126/scitranslmed.3006559. [DOI] [PubMed] [Google Scholar]
- 23.Otto O, Rosendahl P, Mietke A, Golfier S, Herold C, Klaue D, Girardo S, Pagliara S, Ekpenyong A, Jacobi A, Wobus M, Topfner N, Keyser UF, Mansfeld J, Fischer-Friedrich E, Guck J. Nat. Methods. 2015;12:199–202. doi: 10.1038/nmeth.3281. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Lee Y, Jang JH, Li Y, Han X, Yokoi K, Ferrari M, Zhou L, Qin L. Sci. Rep. 2015;5:14272. doi: 10.1038/srep14272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byun S, Son S, Amodei D, Cermak N, Shaw J, Kang JH, Hecht VC, Winslow MM, Jacks T, Mallick P, Manalis SR. Proc. Natl. Acad. Sci. USA. 2013;110:7580. doi: 10.1073/pnas.1218806110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Kai K, Choi DS, Iwamoto T, Nguyen YH, Wong H, Landis MD, Ueno NT, Chang J, Qin L. Proc. Natl. Acad. Sci. USA. 2012;109:18707. doi: 10.1073/pnas.1209893109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yilmaz M, Christofori G. Cancer Metast. Rev. 2009;28:15. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 28.Luster AD, Alon R, von Andrian UH. Nat. Immunol. 2005;6:1182. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Zhang W, Qin L. Angew. Chem. Int. Ed. 2014;53:2344. doi: 10.1002/anie.201309885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Zhou L, Qin L. J. Am. Chem. Soc. 2014;136:15257. doi: 10.1021/ja5072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boneschansker L, Yan J, Wong E, Briscoe DM, Irimia D. Nat. Commun. 2014;5:4787. doi: 10.1038/ncomms5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polacheck WJ, German AE, Mammoto A, Ingber DE, Kamm RD. Proc. Natl. Acad. Sci. USA. 2014;111:2447. doi: 10.1073/pnas.1316848111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.a) Bersini S, Jeon JS, Dubini G, Arrigoni C, Chung S, Charest JL, Moretti M, Kamm RD. Biomaterials. 2014;35:2454. doi: 10.1016/j.biomaterials.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. Proc. Natl. Acad. Sci. USA. 2012;109:13515. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tung CK, Hu L, Fiore AG, Ardon F, Hickman DG, Gilbert RO, Suarez SS, Wu M. Proc. Natl. Acad. Sci. USA. 2015;112:5431. doi: 10.1073/pnas.1500541112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehling M, Frank T, Albayrak C, Tay S. Lab Chip. 2015;15:1276. doi: 10.1039/c4lc01038h. [DOI] [PubMed] [Google Scholar]
- 36.a) Fan Z, Sun Y, Di C, Tay D, Chen W, Deng CX, Fu J. Sci. Rep. 2013;3:2176. doi: 10.1038/srep02176. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Nat. Methods. 2010;7:733. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Reffay M, Parrini MC, Cochet-Escartin O, Ladoux B, Buguin A, Coscoy S, Amblard F, Camonis J, Silberzan P. Nat. Cell Biol. 2014;16:217. doi: 10.1038/ncb2917. [DOI] [PubMed] [Google Scholar]; d) Sun Y, Yong KM, Villa-Diaz LG, Zhang X, Chen W, Philson R, Weng S, Xu H, Krebsbach PH, Fu J. Nat. Mater. 2014;13:599. doi: 10.1038/nmat3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagasse E, Connors H, Al-Dhalimy M, Reitsma M, Dohse M, Osborne L, Wang X, Finegold M, Weissman IL, Grompe M. Nat Med. 2000;6:1229. doi: 10.1038/81326. [DOI] [PubMed] [Google Scholar]
- 38.Herzenberg LA, Parks D, Sahaf B, Perez O, Roederer M, Herzenberg LA. Clin. Chem. 2002;48:1819. [PubMed] [Google Scholar]
- 39.Shields CWt, Reyes CD, Lopez GP. Lab Chip. 2015;15:1230. doi: 10.1039/c4lc01246a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian WY, Zhang Y, Chen WQ. Small. 2015;11:3850. doi: 10.1002/smll.201403658. [DOI] [PubMed] [Google Scholar]
- 41.Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Janicke F, Jackson S, Gornet T, Cristofanilli M, Pantel K. Clin. Cancer Res. 2007;13:920. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 42.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.a) Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc. Natl. Acad. Sci. USA. 2010;107:18392. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang ST, Liu K, Liu JA, Yu ZTF, Xu XW, Zhao LB, Lee T, Lee EK, Reiss J, Lee YK, Chung LWK, Huang JT, Rettig M, Seligson D, Duraiswamy KN, Shen CKF, Tseng HR. Angew. Chem. Int. Ed. 2011;50:3084. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.a) Smith JP, Lannin TB, Syed Y, Santana SM, Kirby BJ. Biomed. Microdevices. 2014;16:143. doi: 10.1007/s10544-013-9814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Deng YL, Zhang Y, Sun S, Wang ZH, Wang MJ, Yu BQ, Czajkowsky DM, Liu BY, Li Y, Wei W, Shi QH. Sci. Rep. 2014;4:7499. doi: 10.1038/srep07499. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sheng WA, Ogunwobi OO, Chen T, Zhang JL, George TJ, Liu C, Fan ZH. Lab Chip. 2014;14:89. doi: 10.1039/c3lc51017d. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zhang JL, Sheng WA, Fan ZH. Chem. Commun. 2014;50:6722. doi: 10.1039/c4cc02002b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Science. 2013;339:580. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng S, Lin HK, Lu B, Williams A, Datar R, Cote RJ, Tai YC. Biomed. Microdevices. 2011;13:203. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.a) Liu ZB, Huang F, Du JH, Shu WL, Feng HT, Xu XP, Chen Y. Biomicrofluidics. 2013;7:011801. doi: 10.1063/1.4774308. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liu ZB, Zhang W, Huang F, Feng HT, Shu WL, Xu XP, Chen Y. Biosens. Bioelectron. 2013;47:113. doi: 10.1016/j.bios.2013.03.017. [DOI] [PubMed] [Google Scholar]; c) Loutherback K, D’Silva J, Liu LY, Wu A, Austin RH, Sturm JC. AIP Adv. 2012;2:42107. doi: 10.1063/1.4758131. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Sun J, Li M, Liu C, Zhang Y, Liu D, Liu W, Hu G, Jiang X. Lab Chip. 2012;12:3952. doi: 10.1039/c2lc40679a. [DOI] [PubMed] [Google Scholar]
- 48.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, Kimura A, Sengupta S, Stott SL, Karabacak NM, Barber TA, Walsh JR, Smith K, Spuhler PS, Sullivan JP, Lee RJ, Ting DT, Luo X, Shaw AT, Bardia A, Sequist LV, Louis DN, Maheswaran S, Kapur R, Haber DA, Toner M. Sci. Transl. Med. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Zeinali M, Hamza B, Engstrom A, Zhu H, Sundaresan TK, Miyamoto DT, Luo X, Bardia A, Wittner BS, Ramaswamy S, Shioda T, Ting DT, Stott SL, Kapur R, Maheswaran S, Haber DA, Toner M. Nat. Methods. 2015;12:685. doi: 10.1038/nmeth.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green BJ, Saberi Safaei T, Mepham A, Labib M, Mohamadi RM, Kelley SO. Angew. Chem. Int. Ed. 2016;55:1252. doi: 10.1002/anie.201505100. [DOI] [PubMed] [Google Scholar]
- 51.Hyun KA, Jung HI. Lab Chip. 2014;14:45. doi: 10.1039/c3lc50582k. [DOI] [PubMed] [Google Scholar]
- 52.Watkins NN, Hassan U, Damhorst G, Ni HK, Vaid A, Rodriguez W, Bashir R. Sci. Transl. Med. 2013;5:214ra170. doi: 10.1126/scitranslmed.3006870. [DOI] [PubMed] [Google Scholar]
- 53.Singh A, Suri S, Lee T, Chilton JM, Cooke MT, Chen W, Fu J, Stice SL, Lu H, McDevitt TC, Garcia AJ. Nat. Methods. 2013;10:438. doi: 10.1038/nmeth.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Almendro V, Marusyk A, Polyak K. Annu. Rev. Phytopathol. 2013;8:277. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
- 55.Gross A, Schoendube J, Zimmermann S, Steeb M, Zengerle R, Koltay P. Int. J. Mol. Sci. 2015;16:16897. doi: 10.3390/ijms160816897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang K, Han X, Li Y, Li SY, Zu Y, Wang Z, Qin L. J. Am. Chem. Soc. 2014;136:10858. doi: 10.1021/ja5053279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, Gerstein M, Snyder M. Nat. Rev. Genet. 2009;10:57. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, Mburu FM, Mantalas GL, Sim S, Clarke MF, Quake SR. Nat. Methods. 2014;11:41. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. Nat. Methods. 2009;6:377. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 60.Streets AM, Zhang X, Cao C, Pang Y, Wu X, Xiong L, Yang L, Fu Y, Zhao L, Tang F, Huang Y. Proc. Natl. Acad. Sci. USA. 2014;111:7048. doi: 10.1073/pnas.1402030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Cell. 2015;161:1187. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Cell. 2015;161:1202. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei W, Shin YS, Ma C, Wang J, Elitas M, Fan R, Heath JR. Genome Med. 2013;5:75. doi: 10.1186/gm479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.a) Lu Y, Chen JJ, Mu LY, Xue Q, Wu Y, Wu PH, Li J, Vortmeyer AO, Miller-Jensen K, Wirtz D, Fan R. Anal. Chem. 2013;85:2548. doi: 10.1021/ac400082e. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lu Y, Xue Q, Eisele MR, Sulistijo ES, Brower K, Han L, Amir ED, Pe’er D, Miller-Jensen K, Fan R. Proc. Natl. Acad. Sci. USA. 2015;112:E607. doi: 10.1073/pnas.1416756112. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ma C, Fan R, Ahmad H, Shi QH, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu CG, Ribas A, Heath JR. Nat. Med. 2011;17:738. doi: 10.1038/nm.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Shi QH, Qin LD, Wei W, Geng F, Fan R, Shin YS, Guo DL, Hood L, Mischel PS, Heath JR. Proc. Natl. Acad. Sci. USA. 2012;109:419. doi: 10.1073/pnas.1110865109. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Xue M, Wei W, Su YP, Kirn J, Shin YS, Mai WX, Nathanson DA, Heath JR. J. Am. Chem. Soc. 2015;137:4066. doi: 10.1021/jacs.5b00944. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Wei W, Shi QH, Remacle F, Qin LD, Shackelford DB, Shin YS, Mischel PS, Levine RD, Heath JR. Proc. Natl. Acad. Sci. USA. 2013;110:E1352. doi: 10.1073/pnas.1303060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang BL, Ghaderi A, Zhou H, Agresti J, Weitz DA, Fink GR, Stephanopoulos G. Nat. Biotechnol. 2014;32:473. doi: 10.1038/nbt.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ng AHC, Chamberlain MD, Situ H, Lee V, Wheeler AR. Nat. Commun. 2015;6:7513. doi: 10.1038/ncomms8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mazutis L, Gilbert J, Ung WL, Weitz DA, Griffiths AD, Heyman JA. Nat. Protoc. 2013;8:870. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang K, Chou CK, Xia X, Hung MC, Qin L. Proc. Natl. Acad. Sci. USA. 2014;111:2948. doi: 10.1073/pnas.1313661111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jo MC, Liu W, Gu L, Dang WW, Qin LD. Proc. Natl. Acad. Sci. USA. 2015;112:9364. doi: 10.1073/pnas.1510328112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skelley AM, Kirak O, Suh H, Jaenisch R, Voldman J. Nat. Methods. 2009;6:147. doi: 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dura B, Dougan SK, Barisa M, Hoehl MM, Lo CT, Ploegh HL, Voldman J. Nat. Commun. 2015;6:5940. doi: 10.1038/ncomms6940. [DOI] [PubMed] [Google Scholar]
- 72.Jang JH, Huang Y, Zheng PL, Jo MC, Bertolet G, Zhu MX, Qin LD, Liu DF. J. Immunol. 2015;195:1320. doi: 10.4049/jimmunol.1403143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khan IU, Serra CA, Anton N, Vandamme T. J. Control. Release. 2013;172:1065. doi: 10.1016/j.jconrel.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 74.a) Liu ZB, Xiao LD, Xu BJ, Zhang Y, Mak AFT, Li Y, Man WY, Yang M. Biomicrofluidics. 2012;6:24111. doi: 10.1063/1.4704522. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Irimia D, Geba DA, Toner M. Anal. Chem. 2006;78:3472. doi: 10.1021/ac0518710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.An F, Qu Y, Liu X, Zhong R, Luo Y. Anal. Chem. Insights. 2015;10:39. doi: 10.4137/ACI.S28905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.a) Nuxoll E. Adv. Drug Deliver. Rev. 2013;65:1611. doi: 10.1016/j.addr.2013.07.003. [DOI] [PubMed] [Google Scholar]; b) Liu ZB, Zhang Y, Yu JJ, Mak AFT, Li Y, Yang M. Sensor. Actuat. B: Chem. 2010;143:776. [Google Scholar]
- 77.a) Han X, Liu Z, Jo MC, Zhang K, Li Y, Zeng Z, Li N, Zu Y, Qin L. Sci. Adv. 2015;1:e1500454. doi: 10.1126/sciadv.1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sharei A, Zoldan J, Adamo A, Sim WY, Cho N, Jackson E, Mao S, Schneider S, Han MJ, Lytton-Jean A, Basto PA, Jhunjhunwala S, Lee J, Heller DA, Kang JW, Hartoularos GC, Kim KS, Anderson DG, Langer R, Jensen KF. Proc. Natl. Acad. Sci. USA. 2013;110:2082. doi: 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Szeto GL, Van Egeren D, Worku H, Sharei A, Alejandro B, Park C, Frew K, Brefo M, Mao S, Heimann M, Langer R, Jensen K, Irvine DJ. Sci. Rep. 2015;5:10276. doi: 10.1038/srep10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whilding LM, Maher J. Mol. Oncol. 2015;9:1994. doi: 10.1016/j.molonc.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.a) Agarwal A, Goss JA, Cho A, McCain ML, Parker KK. Lab Chip. 2013;13:3599. doi: 10.1039/c3lc50350j. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bhatia SN, Ingber DE. Nat. Biotechnol. 2014;32:760. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]; c) Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, Thorneloe KS, McAlexander MA, Ingber DE. Sci. Transl. Med. 2012;4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Science. 2010;328:1662. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Kim HJ, Huh D, Hamilton G, Ingber DE. Lab Chip. 2012;12:2165. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]; f) Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hubner J, Lindner M, Drewell C, Bauer S, Thomas A, Sambo NS, Sonntag F, Lauster R, Marx U. Lab Chip. 2015;15:2688. doi: 10.1039/c5lc00392j. [DOI] [PubMed] [Google Scholar]; g) Torisawa YS, Spina CS, Mammoto T, Mammoto A, Weaver JC, Tat T, Collins JJ, Ingber DE. Nat. Methods. 2014;11:663. doi: 10.1038/nmeth.2938. [DOI] [PubMed] [Google Scholar]; h) Su PJ, Liu Z, Zhang K, Han X, Saito Y, Xia X, Yokoi K, Shen H, Qin L. Sci. Rep. 2015;5:13591. doi: 10.1038/srep13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sung JH, Kam C, Shuler ML. Lab Chip. 2010;10:446. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- 81.a) Griffin DR, Weaver WM, Scumpia PO, Di Carlo D, Segura T. Nat. Mater. 2015;14:737. doi: 10.1038/nmat4294. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang CC, Yang KC, Lin KH, Wu CC, Liu YL, Lin FH, Chen IH. Biotechnol. Bioeng. 2014;111:2338. doi: 10.1002/bit.25295. [DOI] [PubMed] [Google Scholar]
- 82.Robinton DA, Daley GQ. Nature. 2012;481:295. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Polini A, Prodanov L, Bhise NS, Manoharan V, Dokmeci MR, Khademhosseini A. Expert Opin. Drug Discov. 2014;9:335. doi: 10.1517/17460441.2014.886562. [DOI] [PubMed] [Google Scholar]
- 84.Esch EW, Bahinski A, Huh D. Nat. Rev. Drug Discov. 2015;14:248. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Terry SC, Jerman JH, Angell JB. IEEE Trans. Electron Dev. 1979;26:1880. [Google Scholar]
- 86.Nge PN, Rogers CI, Woolley AT. Chem. Rev. 2013;113:2550. doi: 10.1021/cr300337x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, Schuler LA, Alarid ET, Beebe DJ. Lab Chip. 2009;9:2132. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Berthier E, Young EWK, Beebe D. Lab Chip. 2012;12:1224. doi: 10.1039/c2lc20982a. [DOI] [PubMed] [Google Scholar]
- 89.Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, Martel JM, Kojic N, Smith K, Chen PI, Yang J, Hwang H, Morgan B, Trautwein J, Barber TA, Stott SL, Maheswaran S, Kapur R, Haber DA, Toner M. Nat. Protoc. 2014;9:694. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]