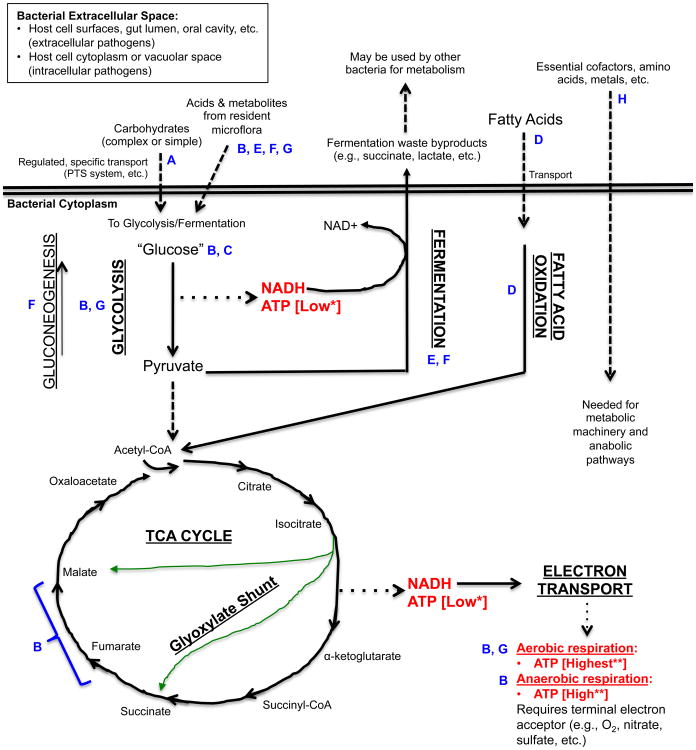
Figure 1. An simplified view of catabolic energy-yielding pathways in bacteria and points of relevance for the indicated bacterial pathogens.
The figure shows a simplified outline of some of the different pathways for utilizing carbohydrates or fatty acids for the generation of ATP. These catabolic pathways and the anabolic pathways that they feed are under extremely complex levels of control, which have been studied mainly in non-infectious organisms (126). However, the unique metabolic strategies employed by infectious bacteria are becoming more appreciated as important aspects of bacterial pathogenesis (1). *ATP generated by substrate level phosphorylation; **ATP generated by oxidative phosphorylation; Solid lines: Metabolic Pathway; Dashed lines: Substrates that feed into catabolic pathways and products generated by bacteria as a result of metabolism or required for metabolism; Dotted lines: Main energy yielding metabolites generated from pathways.
Blue letters indicate specific points of importance for the energy-yielding metabolism of select pathogens listed below (see Table 1 for summary):
A. Pathogenic Streptococci (S. mutans, S. pyogenes, S. pneumoniae); B. Salmonella enterica serovar Typhimurium; C. Brucella abortus; D. Mycobacterium tuberculosis; E. Clostridium difficile; F. Enterohemorrhagic Escherichia coli (EHEC); G. Aggregatibacter actinomycetemcomitans; H. Listeria monocytogenes.