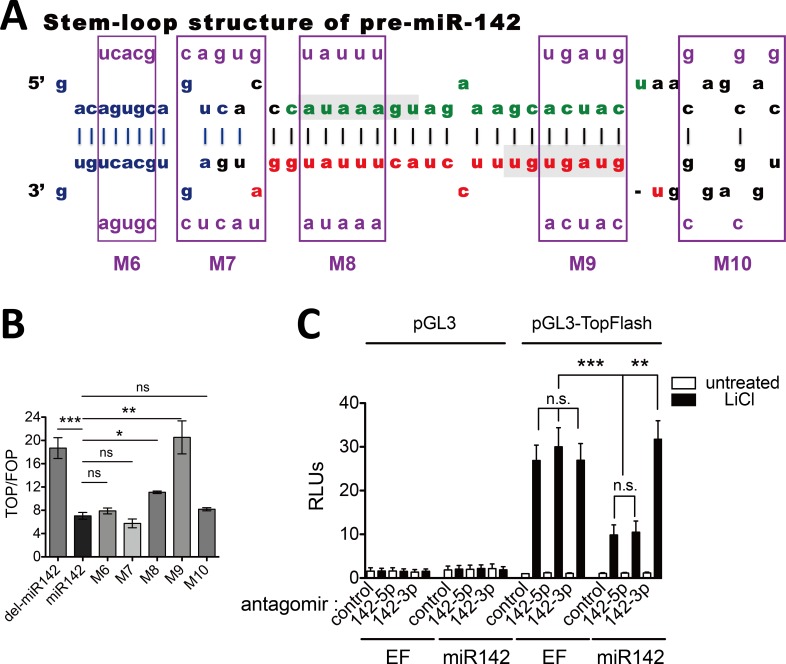
Fig 3. miR-142-3p suppresses Wnt/β-catenin signaling.
(A) Schematic diagram showing the structure of pre-miR-142. The part highlighted in blue indicates sequences surrounding the pre-miR-142. The frames above M6 to M10 mark the positions of five structure-unchanging mutants within or near pre-miR-142, purple bases within the frames indicate the mutant sequences. (B) TopFlash-mediated reporter assay was performed as described in Fig 2D with M6 ~ M10 mutants; error bars mark the SEM (n = 3; ***P < 0.001, **P < 0.01, *P < 0.05, ns P > 0.05, t test). (C) Antagomir-142-3p abolished the effects of miR142. TopFlash mediated reporter assay was performed as in Fig 1C. Antagomir-142-5p, antagomir-142-3p or control antagomir was added, respectively. TopFlash-mediated firefly luciferase activities were normalized to that of Renilla luciferase; error bars mark the SEM (n = 3; ***P < 0.001, **P < 0.01, t test).