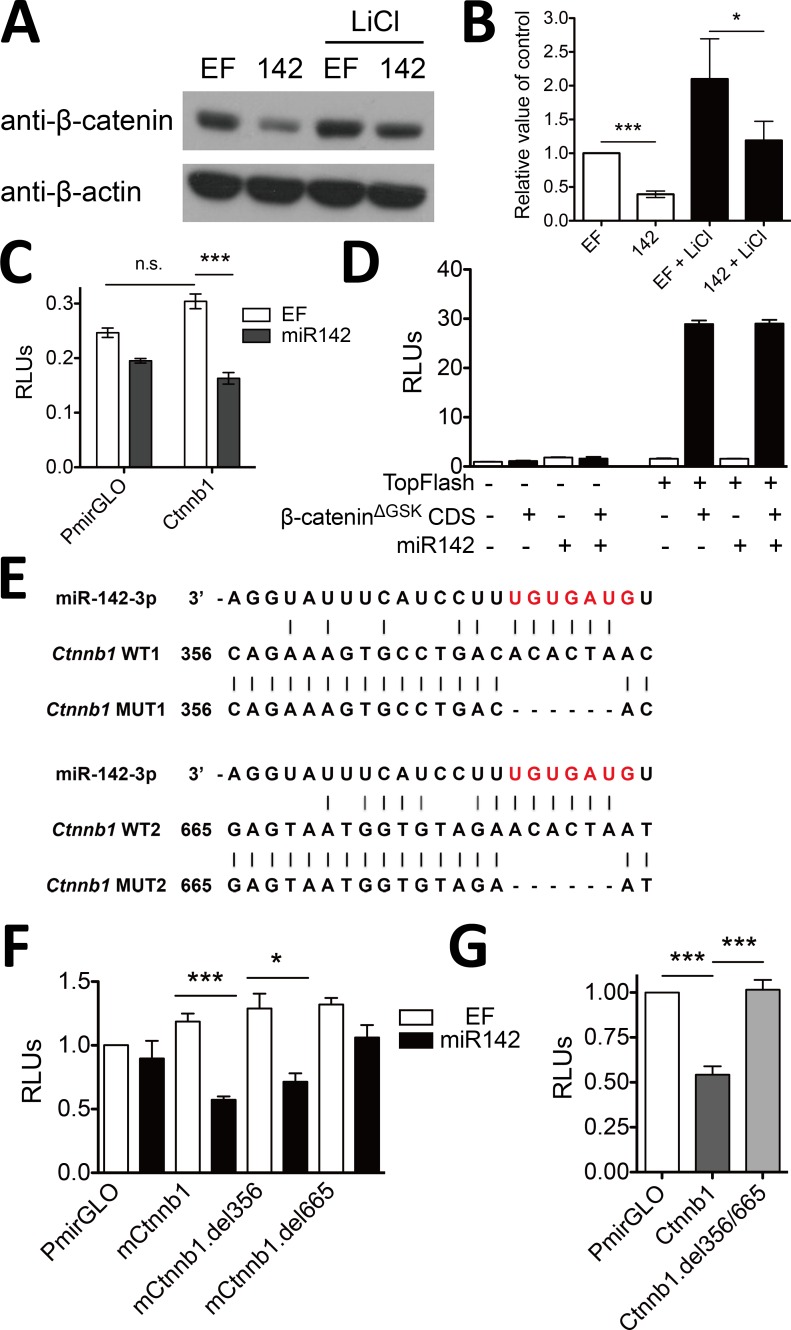
Fig 4. miR-142 directly targets Ctnnb1 mRNA and decreases β-catenin protein level.
(A) Ectopic expression of miR-142 decreased β-catenin protein level. miR-142 expressing vector or empty vector (EF) was transfected into HEK293T cells with or without 25 mM LiCl treatment. (B) Quantitative analysis of the densitometric values of β-catenin signal relative to β-actin; error bars mark the SEM (n = 5; ***P < 0.001, *P < 0.05, t test). (C) miR-142 directly targeted 3’ UTR of β-catenin. Mouse Ctnnb1 3’ UTR was introduced downstream of the luciferase gene in the PmirGLO vector. This vector and miR-142 expressing vector were co-transfected into HEK293T cells. PmirGLO is used as a control. (D) miR-142 did not target CDS of β-catenin. Construct expressing β-cateninΔGSK mutant was co-transfected with pGL3-TopFlash, miR-142 expressing vector, and pRL-TK into HEK293T cells. (E) Schematic representation of two putative miR-142-3p binding sites within mouse Ctnnb1 3’ UTR. Bases highlighted in red indicate the seed sequence of miR-142-3p, and lines in black indicate base pairings. (F) nt665-671 of mouse Ctnnb1 3’ UTR was essential for the regulation by miR-142-3p. PmirGLO or its variant expressing wild type Ctnnb1 3’ UTR or mutated Ctnnb1 3’ UTR (nt356-361- or nt665-671-deletion) was cotransfected with miR-142 expressing vector into HEK293T cells, respectively. (G) Endogenous miR142 targeted to synthetic Ctnnb1 3’ UTR in Jurkat T cells. PmirGLO or its variant expressing wild type Ctnnb1 3’ UTR or mutated Ctnnb1 3’ UTR (nt356-361- and nt665-671-deletion) was electransfected to Jurkat T cells, respectively. Luciferase activities (C, D, F, G) were assessed 24 h after transfection and normalized to the activity of Renilla luciferase; error bars mark the SEM (n = 3; ***P < 0.001, *P < 0.05, t test).