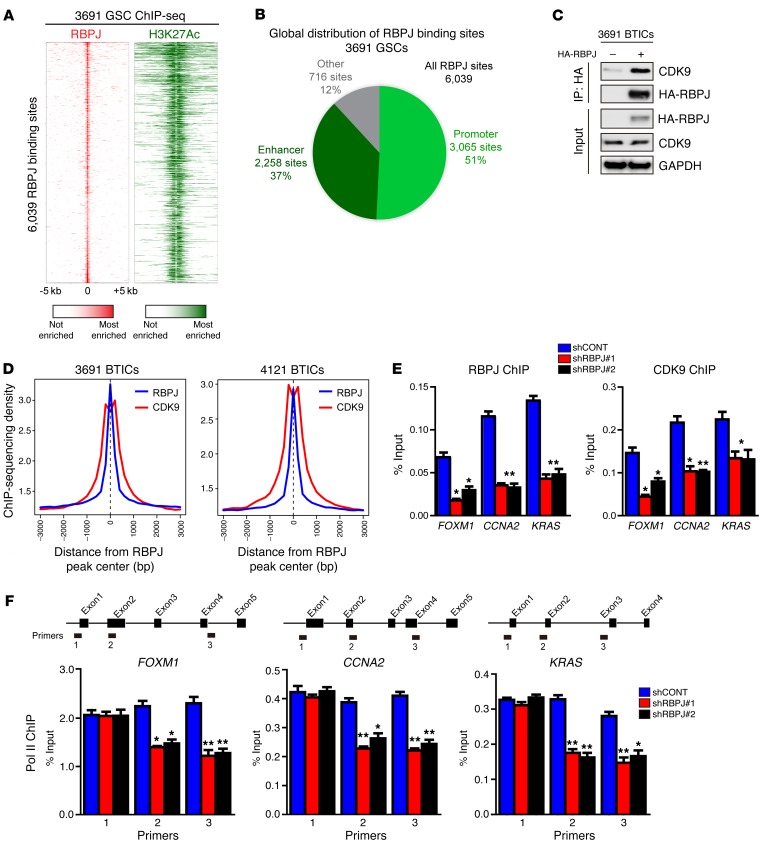
Figure 6. RBPJ binds to CDK9 to promote target gene transcription elongation.
(A) Global analysis of the RBPJ landscape reveals that RBPJ binds nearly exclusively to active promoters and enhancers. ChIP-seq was conducted in 3691 GSCs for both RBPJ and H3K27Ac, a histone mark of active promoters and enhancers. Binding heatmaps of RBPJ (red) and H3K27Ac (green) centered on the center of RBPJ binding sites. Nearly all RBPJ sites are surrounded by H3K27Ac. (B) RBPJ global localization. Active promoters and enhancers were called using H3K27Ac sites. H3K27Ac peaks within 1 kb upstream or downstream of a transcription start site of an expressed gene were considered active promoter sites. All other H3K27Ac sites were considered active enhancer sites. (C) 3691 BTICs cells were transfected with HA-RBPJ plasmid or control vector. Anti-HA immunoprecipitates were immunoblotted with either an anti-CDK9 or anti-HA antibody. Input controls were immunoblotted with indicated antibodies. (D) Aggregate plots of RBPJ and CDK9 ChIP-seq peak intensity centered on RBPJ-bound loci in BTICs. CDK9 ChIP-seq data from GSE51633. (E) Cross-linked chromatin was prepared from 3691 BTICs expressing shCONT, shRBPJ-1, and shRBPJ-2, and then immunoprecipitated with an anti-CDK9 antibody or IgG control, followed by qPCR using primers specific for FOXM1, CCNA2, and KRAS promoters. Knockdown of RBPJ significantly decreased CDK9 recruitment to relevant promoters (2-way ANOVA, *P < 0.05, **P < 0.01, n = 3). (F) Cross-linked chromatin was prepared from 3691 BTICs transduced with shCONT, shRBPJ-1, and shRBPJ-2, and then immunoprecipitated with an anti-POL2 antibody or IgG control followed by qPCR using primers specific for the indicated regions of FOXM1, CCNA2, and KRAS (2-way ANOVA, *P < 0.05, **P < 0.01, n = 3).