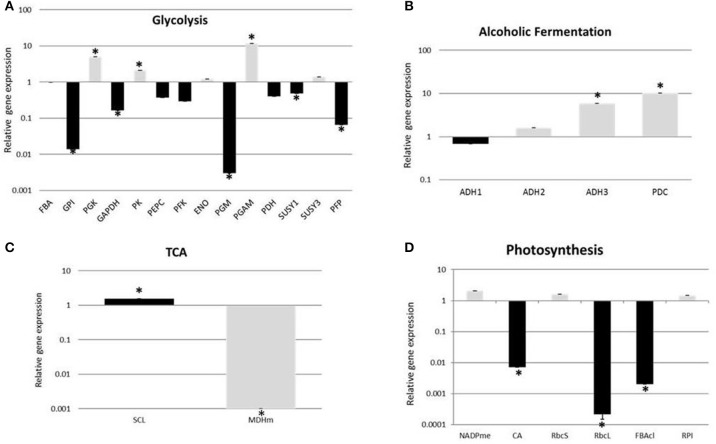
Figure 1.
Comparison of transcripts abundances of genes involved in primary metabolism: (A) glycolysis, (B) Alcoholic fermentation, (C) TCA cycle, and (D) Photosynthesis, in E. grandis cambial zone by RT-qPCR. Data are expressed as log and winter values were used as a control. Expression was determined relative to alpha-tubulin and MDH, as described in Materials and Methods. Asterisks indicates genes that are significantly expressed (P ≤ 0.05). Abbreviations: ALDO, fructose bisphosphate aldolase; GPI, glucose 6 phosphate isomerase; PGK, phosphoglycerate kinase; GAPDH, glyceraldehyde 3 phosphate dehydrogenase; PK, pyruvate kinase; PEPC, phosphoenolpyruvate carboxylase; PFK, ATP-dependent phosphofructokinase; ENO, enolase; PGM, phosphoglucomutase; PGAM, phosphoglycerate mutase; PDH, pyruvate dehydrogenase; SuSy1, sucrose synthase 1; SuSy3, sucrose synthase 3; PFP, PPi-dependent phosphofructokinase; ADH1, alcohol dehydrogenase 1; ADH2, alcohol dehydrogenase 2; ADH3, alcohol dehydrogenase 3; PDC, pyruvate decarboxylase; SCL, succinyl-coa ligase; MDHm, malate dehydrogenase mitochondrial; NADPme, NADP-malic enzyme; CA, carbonic anhydrase; RbcS, rubisco small subunit; RbcL, rubisco large subunit; FBAcl, fructose bisphosphate aldolase chloroplastidial; RPI, ribose-5-phosphateisomerase. Three biological replicates, each with three technical replicates were analyzed per sample and error bars are standard errors of mean.