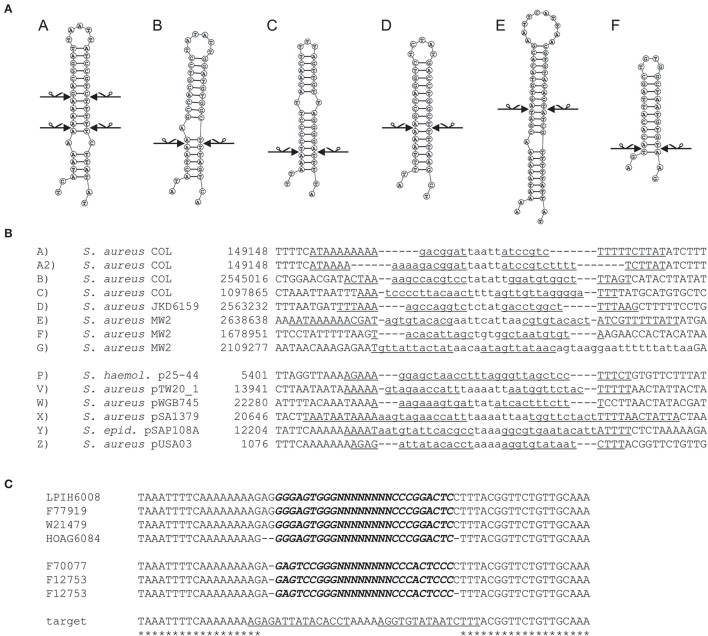
Figure 2.
Insertion sites of TnSha1 in bacterial chromosomes and plasmids. The seven target sites found in the 10 S. aureus strains sequenced in this work have been named from A to G. Staphylococcal plasmid insertion sites are named P to Z. Panel (A) shows the hairpin structures formed by the transposon target sites and the scissors indicate the position of sequence breaks generated upon TnSha1 insertion. Secondary structure predictions were made using RNAstructure web-based software (Reuter and Mathews, 2010). Two independent insertions into target (A) have been identified and both cleavage sites are shown which in each case generate blunt-end structures in the stem of the hairpin. The sequences in both panels (A,B) refer to TnSha1-free target sites in the chromosomes (COL GenBank accession CP000046 and MW2 GenBank accession BA000033) and plasmids. The target sequences in panel (B) report in lower case the sequence deleted upon insertion and the inverted repeats underlined. Panel (C) shows evidence for multiple insertions of TnSha1 in target Z in the repA terminator of plasmids. The aligned sequences show at least four independent insertions in two opposite orientations of TnSha1 into the repA terminator in plasmid pUSA03 (target Z). In this panel TnSha1 sequences are shown in bold italics upper case, the deleted part of the target sequence in lower case, and the repA terminator hairpin underlined. Note that in panel (C) the upper four TnSha1 sequences are in one orientation while the lower three in the opposite orientation. Note that in panel (C) the upper four TnSha1 sequences are in one orientation while the lower three in the opposite orientation. The S. aureus strain names are given on the right.