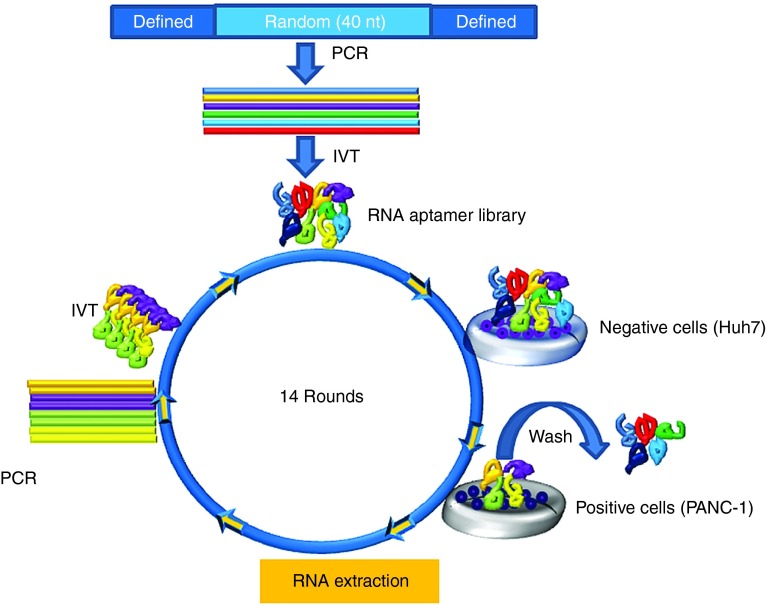
Figure 1.
Naive whole cell-based SELEX. Schematic live-cell SELEX procedures. The DNA library contained 40 nt of random sequences was synthesized and amplified by PCR. 2′F modified RNA aptamer library was synthesized throughout in vitro transcription. To identify the enriched RNA aptamers that bind to target cells, the RNA aptamer library pool was incubated on the negative cells. After removal of nonspecific binding to the negative cells, the supernatant was incubated on the positive cells for positive selection. Total RNA was extracted and amplified through polymerase chain reaction (PCR) and in vitro transcription (IVT). The RNA aptamer selection was repeated for 14 rounds of SELEX. The enriched pools were cloned, and the positive clones were sequenced to identify individual RNA aptamers.