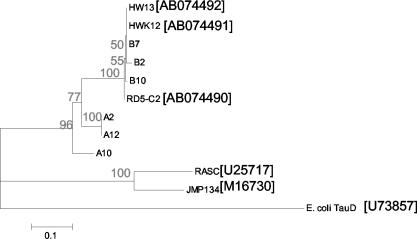
FIG. 6.
Phylogenetic positions of tfdA sequences cloned from Sourhope soil among tfdA sequences from R. eutropha JMP134(pJP4), Burkholderia sp. strain RASC, and reference strains HW12, HW13, and RD2-C5. Strains HW12, HW13, and RD2-C5 are members of the BANA cluster of the α-Proteobacteria most closely related to Bradyrhizobium spp. E. coli tauD encodes taurine/α-ketoglutarate dioxygenase. The neighbor-joining dendrogram (Jukes-Cantor distances) was constructed from reference sequences and common partial sequences (357 bp) of tfdA PCR amplified by using primers tfdAα1 and tfdAα2. Soil clone designations begin with “B ” (for “before 2,4-D application”) or “A ” (for “after 2,4-D application”). Bootstrap confidence limits (percentages) are given at each branch. Scale bar represents a Jukes-Cantor distance of 0.1. GenBank accession numbers are given in brackets after strain designations.