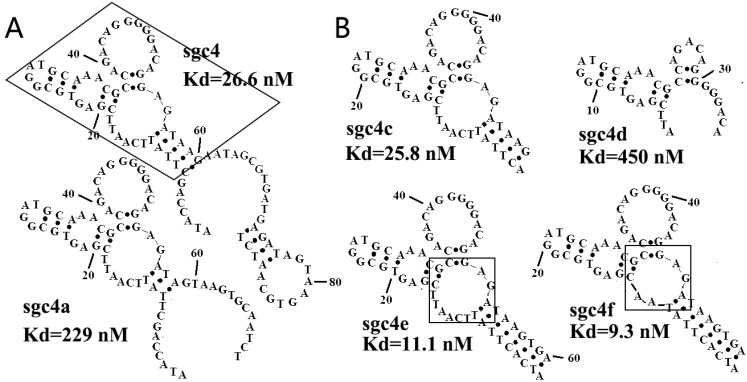
Figure 3.
(A) Secondary structures of sgc4 and sgc4a. Sgc4a had a shorter stem and lower binding affinity than sgc4. Removing nucleotides outside of the region of sgc4 marked with a box resulted in sgc4c, which had a Kd similar to that of full-length sgc4. (B) Truncated and mutated sequences of sgc4, known as sgc4e and sgc4f. It was clear that the long stem in sgc4 and sgc4c was important for maintaining the binding affinity, and that loop marked in box was not the critical binding site (made apparent by the Kd of sgc4f). Sgc4f can be used as the minimal binding motif of sgc4, with 30 fewer nucleotides than sgc4. Reprinted with permission from ref. 46. Copyright (2007) WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.