Abstract
Cytoskeleton-dependent RNA transport and local translation in axons are gaining increased attention as key processes in the maintenance and functioning of neurons. Specific axonal transcripts have been found to play roles in many aspects of axonal physiology including axon guidance, axon survival, axon to soma communication, injury response and regeneration. This axonal transcriptome requires long-range transport that is achieved by motor proteins carrying transcripts as messenger ribonucleoprotein (mRNP) complexes along microtubules. Other than transport, the mRNP complex plays a major role in the generation, maintenance and regulation of the axonal transcriptome. Identification of axonal RNA binding proteins (RBPs) and analyses of the dynamics of their mRNPs are of high interest to the field. Here we describe methods for the study of interactions between RNA and proteins in axons. First, we describe a protocol for identifying binding proteins for an RNA of interest by using RNA affinity chromatography. Subsequently, we discuss immunoprecipitation (IP) methods allowing the dissection of protein- RNA and protein-protein interactions in mRNPs under various physiological conditions.
Introduction
Neurons are highly polarized cells with axons that can reach a length up to 40,000 times longer than their cell body size. While the extremely long axons enable fast transfer of electrical impulses over great distances, they require transport of molecular complexes in order to be sustained. This long distance transport is achieved by movement of motor proteins on microtubules in the axons. Since transport over axonal distances requires significant investments of both time and energy, targeting of mRNAs to axons allows local synthesis of proteins upon need. For instance, local translation of β-actin and Par3 mRNA was reported in netrin or BDNF-induced attractive turning of neuronal growth cones (Hengst, Deglincerti, Kim, Jeon, & Jaffrey, 2009; Leung et al., 2006; Yao, Sasaki, Wen, Bassell, & Zheng, 2006). Another example comes from injured axons of mature neurons where local translation of importin β1 can facilitate the cell’s response to injury and subsequent regeneration (Perry et al., 2012; Rishal & Fainzilber, 2014). Increasing evidence supports the importance of axonal RNA translation in multiple aspects of axon biology including guidance (Piper et al., 2006; K. Y. Wu et al., 2005), survival (Cox, Hengst, Gurskaya, Lukyanov, & Jaffrey, 2008; Yoon et al., 2012), injury response (Ben-Yaakov et al., 2012; Hanz et al., 2003; Perlson et al., 2005; Yudin et al., 2008) and regeneration (Verma et al., 2005; Zheng et al., 2001).
RNA is transported to axons in the form of granules: it interacts with RNA binding proteins (RBPs), adaptor and motor proteins to form an mRNA-protein complex (Fritzsche et al., 2013). RBPs play a crucial role in the regulation of RNA localization and local translation, as they act as transporters as well as inhibitors or initiators of translation in response to specific cues (Sasaki et al., 2010). Therefore, studying the specific RBPs that are associated with axonal mRNAs of interest is valuable to the understanding of axonal local translation. The specificity of the interaction between RBPs and mRNA is determined by recognition of either primary or secondary structures in the RNA molecule, such as stem-loop or G-quartet structures (Anko & Neugebauer, 2012; Aviv, Lin, Ben-Ari, Smibert, & Sicheri, 2006; Darnell et al., 2001). Thus, identifying elements in the RNA sequence and structure is important to understand its interactions with various RBPs.
A number of RBPs have been identified in axons so far, supporting their role in regulating mRNA transport and translation (Gomes, Merianda, Lee, Yoo, & Twiss, 2014). Multiple techniques, such as RNA immunoprecipitation (RIP) and cross-linking immunoprecipitation (CLIP) have been developed in order to identify their associated mRNAs (Jensen & Darnell, 2008; Licatalosi et al., 2008; Milek, Wyler, & Landthaler, 2012; Niranjanakumari, Lasda, Brazas, & Garcia-Blanco, 2002). However, these approaches focus on profiling the mRNAs that bind to a target RBP, but are not useful if the relevant RBP is not known. Other approaches have been developed to identify RBPs that bind to specific target RNA. For example, affinity purification using aptamer-tagged mRNAs such as the MS2 and PP7 systems and subsequent identification of bound RBPs (Hogg & Goff, 2010; Slobodin & Gerst, 2010). The recently developed CRISPR/Cas technique also provides a method to specifically purify RNA-RBP complexes (O’Connell et al., 2014). While these techniques are powerful in purifying RBPs associated to a target RNA, they require transfection/delivery of exogenous RNA and accessory proteins (e.g., MS2 coat protein, catalytically inactive Cas9) and thus, are impractical for detecting RNA-RBP interaction in axons in vivo. Another approach is to use complementary cDNA or RNA aptamers in order to pull down target mRNA along with their associated RBPs for further detection (Castello et al., 2012; Leppek & Stoecklin, 2014; Lingner & Cech, 1996). While this technique doesn’t suffer from the caveat mentioned above, its yield is limited by poor accessibility of the target sequence inside the mRNP tightly packed complex and thus is not compatible with large-scale proteomics.
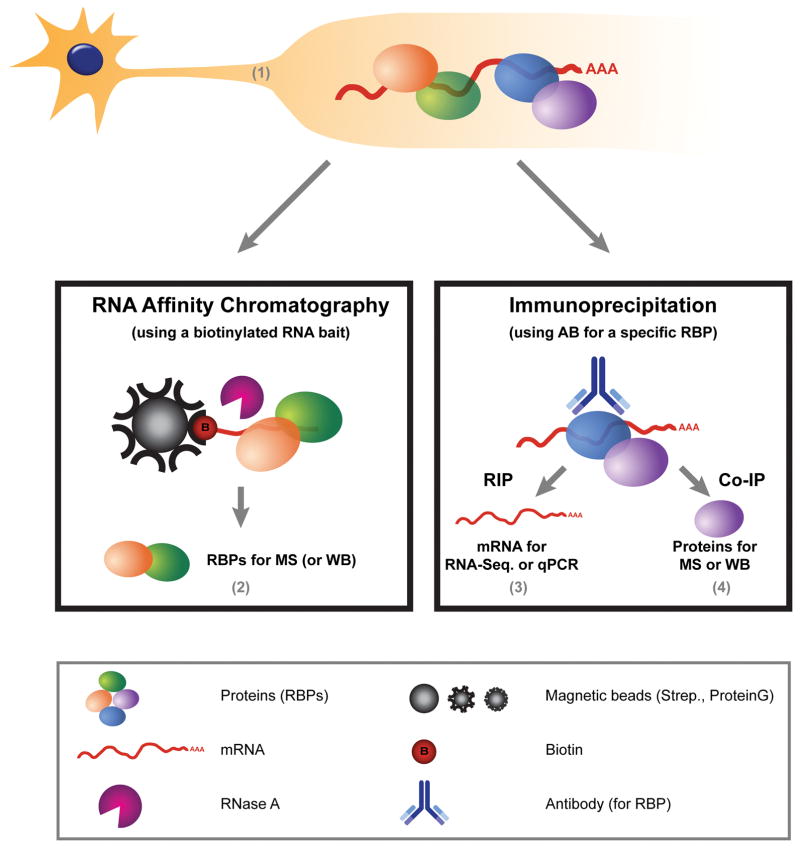
Here we describe a method for RNA affinity chromatography from sciatic nerve axoplasm under native conditions, enabling identification of RBPs by subsequent mass spectrometry. We further describe a complementary approach to verify the RBP-RNA interaction with RIP and study the effects of different conditions on the composition of the mRNP by RIP and co- immunoprecipitation (co-IP) of proteins (see Figure 1).
Figure 1. Characterization of mRNA-Protein complexes in axons of mature neurons.
Overview of the different methods described in this chapter. Axonal material, containing mRNP complexes, is isolated as (described in section 1). Later, RNA affinity chromatography is performed in order to identify novel RBPs that bind to a specific RNA-bait (Section 2). Further, we describe a complementary approach, using an antibody (AB) for a specific RBP, to verify the RBP-RNA interaction (3, RIP) or protein-protein interactions (4, Co-IP) within the axonal mRNP complex.
Methods
1. Preparation of axoplasm enriched in axonal proteins from sciatic nerves
1.1. Materials
Nuclear transport buffer: 20 mM HEPES (pH 7.3), 110 mM potassium acetate, 5 mM magnesium acetate, supplemented with protease/phosphatase/RNase inhibitors
Micropestles
1.2. Procedure
Dissect sciatic nerves directly into nuclear transport buffer (1 sciatic nerve in 50/100 μl buffer for mouse/rat, respectively). Squeeze tissue manually with a pellet micropestle suitable for an eppendorf tube, until the tissue loses its fibrous consistency. Centrifuge 10,000 g for 10 minutes at 4°C. Discard pellet and continue with supernatant.
Remarks
** Total axoplasm quantity should be adjusted according to the abundance of the interactions of interest. In our hands, 8–10 mouse sciatic nerves (300–400 μg) pooled in 400 μl nuclear transport buffer are enough for subsequent pull downs (whether RNA is extracted for quantitative PCR or proteins are extracted for Western blot analysis). Alternatively, 2–4 rat sciatic nerves (200–400 μg) can be used. For high throughput methods such as mass spectrometry and RNA sequencing, quantities should be increased as necessary (we recommend starting with 600–800 μg of protein per sample).
** The axoplasm protocol contains no detergent so that non-neuronal cells in the nerve should not be lysed. If detergent is needed for lysis (such as in the case of extraction from whole cells), NP-40 can be added at a concentration of 0.1%. This amount of detergent will not affect native RNA-protein and protein-protein interactions. If detergent is needed when working with axoplasm, it should be added after the centrifugation step so that glial cells are not lysed in order to retain enrichment for axonal proteins.
2. RNA affinity chromatography
This protocol is used in order to identify binding proteins for a known target RNA. Whether the full transcript is to be queried or only a known fragment (such as a localization element or a regulatory structure), bait is produced by attaching biotin to the RNA molecule. In order to study RBPs that are unique to or of special importance in axons in particular, we recommend using baits that are the minimal known mRNA motif that is critical for conferring axonal localization of the transcript of interest (if known). Examples for such motifs are the differential sequences between the long and short forms of the 3′UTR of importin β1 (Perry et al., 2012) and Impa1 (Andreassi et al., 2010), that were shown to contain axonal localization elements.
Biotinylated RNA baits immobilized on streptavidin-coated beads are incubated with the axoplasm or tissue lysate of interest. The bound proteins are subsequently eluted for analysis (Figure 2) by proteomics and mass spectrometry approaches, or alternatively processed for Western blot (WB) analysis (for the verification of individual interactions).
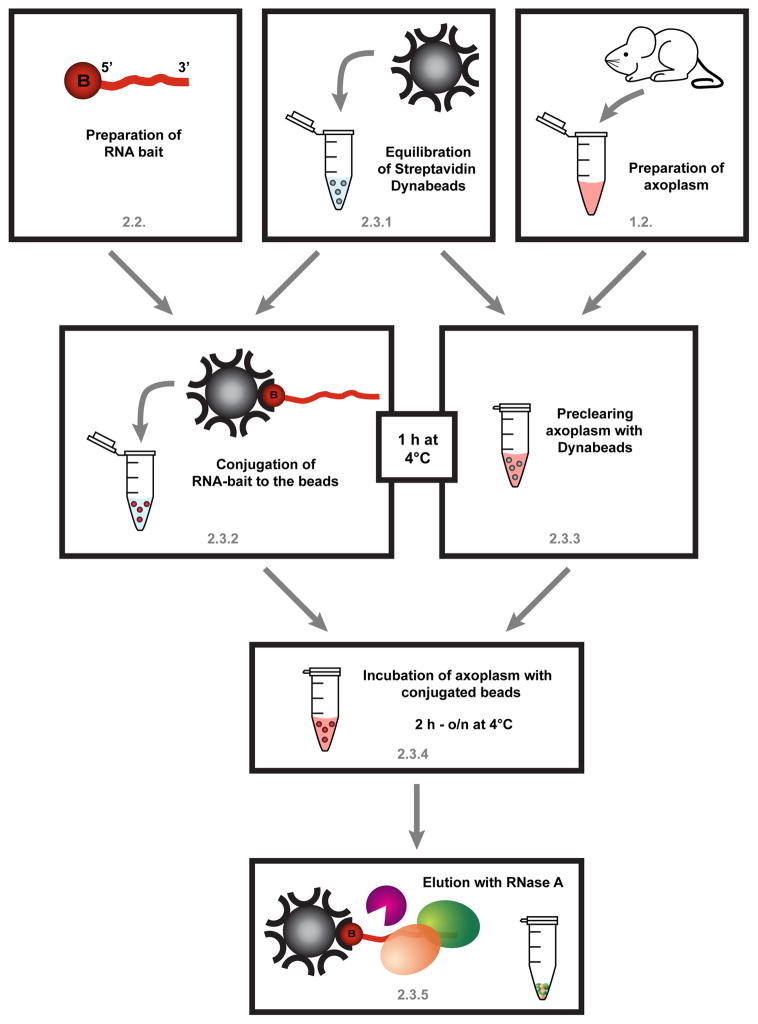
Figure 2. Schematic workflow for RNA affinity chromatography, described in section 2.
First, the biotinylated RNA probe is prepared (section 2.2), preferably in advance. However the axonal extract should be prepared fresh each time (section 1.2.). The last centrifugation step of the axoplasm preparation can be used to equilibrate Streptavidin Dynabeads. Half of the beads will be used for the preclearing of the axoplasm (2.3.3), the other half will be linked to the RNA bait (2.3.2). Since both steps require incubation for 1h at 4°C, this can be performed in parallel. Afterwards, the axoplasm will be transferred to the conjugated beads and incubated for at least 2h to o/n at 4°C with overhead rotation (2.3.4). After 3 washes in high salt buffer proteins are eluted using RNase A and the sample is prepared for MS (2.3.5).
2.1. Materials
T7 RNA Polymerase (Roche, 10881767001)
SP6 RNA Polymerase (Roche, 10810274001)
Biotin RNA labeling mix (Roche, 11685597910)
DNase I (Roche, 04716728001)
Mini Quick Spin column (Roche, 11814397001)
Buffer A: 100 mM NaOH, 50 mM NaCl
Buffer B: 50 mM NaCl
2x Binding buffer: 10 mM Tris-HCl (pH 7.5), 2 M NaCl, 1 mM EDTA
1x Binding buffer: 5 mM Tris-HCl (pH 7.5), 1 M NaCl, 0.5 mM EDTA
Low salt wash buffer: 10 mM HEPES (pH 7.4), 3 mM MgCl2, 14 mM NaCl, 1 mM DTT and 5% glycerol
High salt wash buffer: 10 mM HEPES (pH 7.4), 3 mM MgCl2, 250 mM NaCl, 1 mM DTT and 5% glycerol
Elution buffer: mix 380 μl of high salt wash buffer with 20 μl of RNase A (10 mg/ml, Sigma R4875)
Streptavidin Magnetic Dynabeads (Invitrogen, 11205D)
Vertical rotator for eppendorf tubes (“overhead rotation”)
2.2. Preparation of RNA bait
RNA affinity chromatography is based on working with a known RNA of interest. If a specific sequence element of up to 100 nucleotides is to be queried, it is recommended to use a synthetic 5′ end biotin labeled RNA oligonucleotide synthesized to order. If the full transcript or an element larger than 100 nucleotides is to be queried, bait should be prepared via in vitro transcription with biotin-UTP using the following protocol. We recommend preparing the biotinylated RNA bait in advance.
2.2.1 In vitro transcription with biotin-UTP
Clone the DNA template for the RNA bait into plasmids with T7 or SP6 promoters
Linearize plasmid by using restriction endonuclease
-
Prepare an in vitro transcription reaction in PCR tubes as below:
10x Biotin RNA Labeling mix 2 μl 10x Transcription buffer 2 μl RNA polymerase (SP6 or T7) 2 μl 1 μg of linearized plasmid DNA + nuclease free water 14 μl Incubate at 37°C (T7) or 42°C (SP6) for 2 h
Add 2 μl of DNaseI and incubate for 15 min at 37°C
Add 0.8 μl of 0.5 M EDTA to stop the reaction
Add nuclease free water up to 100 μl
Use spin column to remove unincorporated nucleotides
Run a sample on an agarose gel to check for proper size of synthesized RNA bait
Determine RNA concentration by Nanodrop or Ribogreen (Invitrogen)
Adjust concentration to 0.1 mM with nuclease free water
Aliquot and store at −80°C
2.2.2 Synthesis of biotinylated RNA oligonucleotide
Biotinylated RNA oligonucleotides can be purchased commercially. Biotin should be attached to the 5′-terminus of the RNA oligonucleotide. In our experience, up to 100 nucleotides length synthetic RNA oligonucleotides provide a sufficient substrate for RBP interactions. However, shorter oligonucleotides will incur fewer costs and may provide higher specificity interactions. The oligo should be reconstituted to 0.1 mM concentration in nuclease free water.
Remarks
** Appropriate negative controls should be included. Free biotin (no RNA probe) can serve as a general control. Additionally, if information is available on potential binding motifs within the RNA bait, it is recommended to include control baits with suitably mutated sequences. Alternatively, the complementary sequence of the target RNA can be used as a control, since it will have identical GC ratio. However, care should be exercised as the complementary strand may in part assume a similar stem-loop structure as the target RNA and might be recognized by similar RBPs.
2.3. RNA affinity chromatography
2.3.1 Equilibration of Streptavidin-Dynabeads
For each pull down, use 50 μl of Streptavidin-Dynabeads. Prepare additional 50 μl beads per sample for preclearing the axoplasm sample.
Wash beads with 1 ml of buffer A
Repeat step 1
Wash beads with 1 ml of buffer B
Wash beads with 1 ml of 2x binding buffer
Resuspend beads in 0.4 ml of 1x binding buffer
2.3.2 Conjugation of RNA bait to the beads
Add 1 μl of RNase inhibitor (Promega, 40 unit/μl) into resuspended beads
Add 10 μl of 100 ng/μl in vitro transcribed RNA (approximately 3 μM of 100 nucleotide RNA) or 4 μl of 0.1 mM synthetic RNA
Incubate for 1 h with overhead rotation at 4°C
Wash RNA-bound dynabeads with 1x binding buffer three times to remove unbound RNA
Completely remove 1x binding buffer and rinse beads with low salt buffer two times
2.3.3 Preclearing axoplasm with Dynabeads
Wash beads from step 2.3.1.5 three times with low salt wash buffer
Add axoplasm and incubate for 30–60 min at 4°C with overhead rotation
2.3.4 Incubation of cleared axoplasm with RNA-bound beads
Add precleared axoplasm into RNA bound beads (from step 2.3.2.5) and supplement with RNase inhibitor to a final concentration of 200 units/ml
Incubate 3 h at 4°C with overhead rotation
2.3.5 Washing and elution
Wash beads with 1 ml of high salt wash buffer 5–6 times (supernatant can be extracted for processing if desired)
After the last wash, centrifuge tubes and re-place on the magnet in order to remove all traces of the wash buffer
Add 30 μl of elution solution and incubate at 37°C for 30 min with occasional mixing
Centrifuge with maximum speed, re-place tubes on magnet and transfer the supernatant solution into new tubes
Add 10 μl of 4x Laemmli buffer, boil and run SDS-PAGE followed by mass spectrometry (MS) or Western blot analysis
Remarks
** In this protocol, we describe elution using RNase A. The advantage of this method is that it specifically elutes the proteins that are precipitated through interaction with the RNA, rather than directly with the beads. Different elution methods can be utilized, each with different specificity and subsequent different yields. We have attempted elution with Laemmli buffer (30 μl of 1x buffer incubated 10 minutes at 65°C), which resulted in better yields than the aforementioned RNase A elution. However this also resulted in increased elution of non- specifically bound components from the beads, increasing background in subsequent mass spectrometry analyses. Another approach is to use photo-cleavable biotin and elute via UV radiation. For our purposes this rarely produced enough material for analyses.
** RNase A is a ribonuclease that cleaves single strand RNA. Therefore, when designing synthetic RNA oligo, take into consideration that the bait RNA molecule should have a stretch of at least 10 nucleotides that are predicted to be single stranded and not folded into secondary structures.
** If the immunoprecipitated proteins are to be sent for MS, it should be considered that the RNase A used for elution is present in excess to the sample and therefore, could interfere with MS analysis. In order to overcome this issue we recommend preparing the proteins for MS by on-gel digestion rather than directly on-beads digestion. This is achieved by running SDS-PAGE, staining the gel with MS-compatible reagents (of appropriate sensitivity for protein levels) and conducting protein-digestion from the gel pieces, excluding the piece containing the prominent RNase A band. By doing so, some of the RNase A should be separated by virtue of its size thus relieving the sample from the excessed amount.
3. RNA immunoprecipitation
This protocol is used to isolate the mRNP complex by pulling down a specific RBP with an antibody and then extracting the associated mRNAs. Validation of interactions with specific transcripts (such as those identified with the help of the protocols described in Section 2, RNA affinity chromatography) can be done by subsequent quantitative PCR (qPCR). Alternatively, eluted RNA can be processed for high throughput RNA sequencing to identify novel transcripts interacting with the queried RBP. Here, we suggest an indirect method for pull down by incubating the antibody with the axonal extract for several hours prior to adding the beads (see Figure 3 for workflow). We have found that this can increase yields of RNA as compared to protocols requiring prior coupling of antibody to the beads, perhaps due to steric hindrance of mRNP recognition by immobilised antibody.
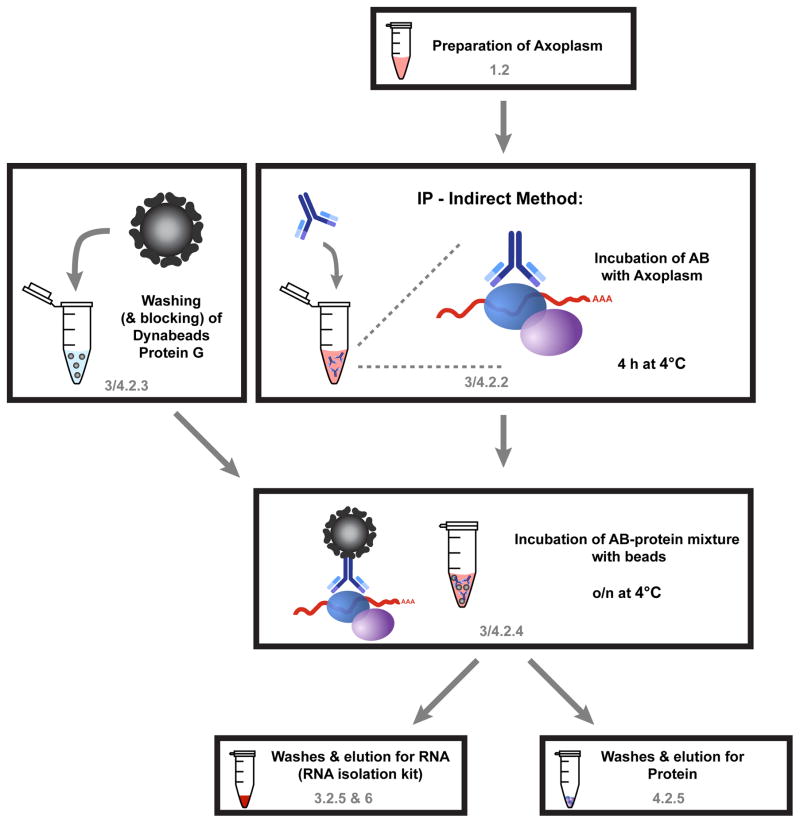
Figure 3. Schematic workflow for RIP and Co-IP as described in section 3 and 4, respectively.
The first steps of the protocol are the same for RIP (section 3) and Co-IP for proteins (section 4). Only the final washing steps and the elution procedure differ for the two protocols. Axoplasm is prepared as described in 1.2 (notice modifications for section 3) and incubated with the RBP-antibody for 4h at 4°C (3.2.2). During this incubation period, protein G magnetic beads are washed (and we recommend blocking for 1h) so that after 4h the axoplasm-AB mixture can be added to the beads. After incubation over night at 4°C the beads are washed and either RNA (3.2.5 & 6) or proteins (4.2.5) are eluted.
3.1. Materials
Wash buffer 1: Nuclear transport buffer supplemented with 1% NP-40
Wash buffer 2: High salt buffer: 50 mM Tris-HCl (pH 7.5), 300 mM KCl, 12 mM MgCl2, 1% NP-40, 1 mM DTT supplemented with Complete Protease inhibitor- EDTA free (Roche), RNase Inhibitor (Promega, 200 U/ml), Ribonucleoside-vanadyl complex (Sigma-aldrich)
1x PBS
PBS-T: 1x PBS supplemented with 0.02% Tween-20 (Sigma-aldrich)
Salmon sperm DNA (Invitrogen, 15632-011)
Protein G dynabeads (Invitrogen, 10004D)
Vertical rotator for eppendorf tubes (“overhead rotation”)
Remarks
** Since the desired outcome of this protocol is gaining good quality RNA for subsequent analyses, special care should be taken in order to avoid RNA degradation by RNases. All solutions should be made with nuclease-free water and supplemented with fresh RNase inhibitors, as indicated, before use. Tubes should be RNase free and all working surfaces and tools should be thoroughly cleaned with an RNase decontamination solution (such as RNaseZap, Life technologies, AM9870). We highly recommend separating the work area for this protocol from that used for the RNase treatment step in section 2.3.5, washing and elution of the RNA affinity chromatography protocol.
3.2. RNA immunoprecipitation
3.2.1 Preparation of axoplasm
Axoplasm is prepared according to the protocol described in section 1, Preparation of axoplasm enriched in axonal proteins from sciatic nerves. After centrifugation, supernatant is transferred to a new tube and NP-40 is added to the axoplasm to a final concentration of 1%.
Take 10% of the extract as ‘input’ for subsequent analyses.
3.2.2 Incubation of antibody with protein extract
Add 10 μg of antibody to axoplasm
Incubate 4 h at 4°C with overhead rotation
Remarks
** Suitable negative controls should be employed. In our hands, the most specific control was to incubate the antibody with the immunization peptide used to produce it prior to incubation with axoplasm. For 10 μg antibody, add 50 μg peptide or equivalent volume of nuclease free water and incubate for 1 h at room temperature with rotation of 45° at 20 RPM. Add the whole mixture into axoplasm at step 3.2.2. If an epitope-mimicking peptide is not available, perform comparative immunoprecipitations with equivalent amounts of axoplasm and total IgG from a non-immunized animal of the same host species.
** Different antibodies have different affinities and will need to be individually optimized; therefore, for some antibodies concentrations different then 10 μg per sample may be better. Adjust the concentration according to your specific antibody.
3.2.3 Preparation of protein G beads
Take 100 μl beads per sample
Wash beads in 1 ml PBS-T
Wash beads twice with wash buffer 1
Suspend beads in 1 ml wash buffer 1 then add 8 μg salmon sperm DNA per 100 μl beads to block non-specific interactions to beads
Incubate 1 h at 4°C with overhead rotation
Wash three times in wash buffer 1
Distribute into protein low-bind eppendorf tubes (1 tube per sample)
3.2.4 Incubation of antibody-protein mixture with beads
Extract wash buffer from beads
Transfer antibody-extract mixture from step 3.2.2.2 to the beads
Incubate overnight at 4°C with overhead rotation
3.2.5 Washing and elution
Collect supernatants in pre-labeled tubes
Wash beads three times with 600 ml wash buffer 2 for 5 min at 4°C with overhead rotation
3.2.6 RNA extraction
Various RNA extraction methods can be utilized. In our hands, using kits designed for RNA extraction from micro-sized samples worked better than common chloroform phase-separation methods. The use of kits may compromise yields as compared to phase-separation, however, samples are generally cleaner and thus more compatible with downstream applications. We recommend using the RNAqueous-Micro total RNA isolation kit (Life technologies, AM1931) with modifications as follows:
Elute RNA from beads by applying 200 μl of lysis buffer on beads
Vortex on low speed 5 times for 4 seconds
Extract buffer from beads into a fresh RNase-free tube
Add 100 μl 100% ethanol then pipette up and down to mix solution
Load sample onto columns and wash as described in kit protocol
Elute with 8 μl elution buffer (supplied with the kit)- Repeat elution twice
Remarks
** To DNase or not to DNase? Usually, it is recommended to treat RNA samples with DNase I after extraction in order to get rid of DNA traces that can interfere with downstream applications. However, DNase treatment can lead to loss of RNA as well as DNA. In our hands, DNase treatment had a detrimental effect on RNA yields. Therefore, depending on the downstream application, if DNase treatment can be avoided we highly recommend to skip it. If the downstream application is qPCR, the lack of DNase treatment can be compensated for by working with primers that span exon-exon junctions and thus should not recognize genomic DNA. In addition, some cDNA transcription kits have a milder DNase treatment step that might minimize losses of RNA.
3.3. Evaluation of eluted RNA by qPCR
If a specific interaction is to be validated through the RIP method, evaluating the amounts of a specific transcript in the elution can be achieved by quantitative PCR (real time PCR or digital droplet PCR).
3.3.1 cDNA synthesis
As in the case for RNA extraction, many commercial kits are available for first strand cDNA synthesis. Two important considerations should be taken into account when choosing a suitable kit:
Dynamic range of the transcriptase: As different mRNAs have different abundances in the eluted RNA, and also different lengths, it is important to work with a transcriptase that will not introduce biases towards less or more abundant transcripts, as well as biases towards shorter or longer transcripts.
Choice of primers: cDNA synthesis kits commonly offer the choice between random hexamers or oligo-dT primers. In our hands, random hexamers gave the most consistent results with RNA pulled down by RBPs. This might be due to the fact that some transcripts will not elute as a whole, but rather only as fragments. Alternatively, some kits utilize both oligo-dT and random hexamers in an attempt to reduce 5′ and 3′ biases.
3.3.2 Quantitative PCR reaction
Detection and quantification of specific transcripts can be done by standard qPCR. Depending on the abundance of a specific transcript, either double-stranded DNA-binding dyes (e.g. SYBR Green) can be used - or fluorescent probes (e.g., TaqMan) which require higher concentration but are also more specific. Typically, RIP will result in low quantities of RNA and therefore SYBR Green will be the most compatible choice. Relative quantification should be performed by measuring the level of the immunoprecipitated RNA versus input to derive the degree of enrichment of the transcript in the RBP immunoprecipitates.
4. Co-immunoprecipitation to identify interacting RBPs and motor proteins
This protocol is complementary to the RIP described in section 3. It is used to isolate an mRNP complex by immunoprecipitating a specific RBP, followed by analyses of associated proteins. This can be done in order to study changes in the protein components of the mRNP following various treatments and in different physiological conditions. In addition, this protocol can be used in order to determine if the axonal RBP interacts with molecular motors (i.e., kinesin, dynein, or myosin) responsible for its transport into and within axons. The protocol is very similar to the RIP protocol described in section 3, with minor changes in buffers and washes to better preserve protein-protein interactions (Figure 3).
4.1. Materials
Nuclear transport buffer, supplemented with protease, phosphatase and RNase inhibitors as mentioned before
Wash buffer 1: Nuclear transport buffer, supplemented with 0.1% NP-40
Wash buffer 2: Nuclear transport buffer, supplemented with 0.5% NP-40
Wash buffer 3: Nuclear transport buffer, supplemented with 1% NP-40
1x PBS
PBS-T: 1x PBS supplemented with 0.02% tween-20
Salmon sperm DNA (Invitrogen, 15632-011)
Protein G Dynabeads (Invitrogen, 10004D)
Vertical rotator for eppendorf tubes (“overhead rotation”)
4.2. Co-immunoprecipitation
4.2.1 Preparation of axoplasm
Axoplasm is prepared according to the protocol described in section 1, without addition of NP-40.
Reserve 10% of the extract as an ‘input’ sample for subsequent analyses.
4.2.2 Incubation of antibody with protein extract
Add 10 μg of specific antibody or control (see section 3.2.2 remarks) to axoplasm
Incubate 4 h at 4°C with overhead rotation
4.2.3 Preparation of protein G beads
Take 100 μl beads per sample
Wash with 1 ml of PBS-T
Wash beads twice in nuclear transport buffer
Suspend beads in 1 ml wash buffer 1 then add 8 μg salmon sperm DNA per 100 μl beads to block non-specific interactions to beads
Incubate 1 h at 4°C with overhead rotation
Wash three times in nuclear transport buffer
Distribute into protein low-bind eppendorf tubes (1 tube per sample)
4.2.4 Incubation of antibody-protein mixture with beads
Extract buffer from beads
Transfer antibody-extract mixture from step 4.2.2.2 to the beads
Incubate overnight at 4°C with overhead rotation
4.2.5 Washing and elution
Collect supernatants in pre-labeled tubes
Wash with 600 μl nuclear transport buffer for 5 min at 4°C with overhead rotation
Wash with 600 μl wash buffer 1 for 5 min at 4°C with overhead rotation
Wash with 600 μl wash buffer 2 for 3 min at 4°C with overhead rotation
Wash with 200 μl wash buffer 3 for 2 min at 4°C with overhead rotation
Wash with 600 μl nuclear transport buffer for 5 min at 4°C with overhead rotation
Resuspend in 600 μl 1x PBS and transfer the beads to a new labeled tube
Remove PBS and apply 40 μl of 1x Laemmli buffer
Elute by incubating 10 minutes at 65°C with occasional mixing
Extract supernatants to a new tube, discard beads
Run SDS-PAGE followed by immunoblotting to confirm candidate interactions or alternatively, conduct mass spectrometry in order to screen for novel interactors
Remarks
** If co-immunoprecipitated proteins are to be sent for mass spectrometry (MS), it should be considered that the bait antibody is present in excess to the sample and therefore, could interfere with MS analysis. In order to overcome this issue the ‘direct’ approach for immunoprecipitation can be utilized, where the antibody is first coupled to the beads and covalently crosslinked so it is not eluted into the sample at later stages. (We have successfully used the amine-to-amine crosslinker bis(sulfosuccinimidyl) suberate (BS3). Sigma-aldrich, S5799). In case the RBP of interest is of low abundance and the indirect method is preferable, efforts should be made to deplete antibody from the eluted sample before MS.
Discussion
Here, we describe methods to study native mRNPs containing a transcript of interest in axons of peripheral nerves under different physiological conditions. First, we identify RBPs specific for a known mRNA by affinity chromatography with the RNA as bait. After identification, target RBPs are subjected to both co-IP and RIP in order to study the changes in the mRNP complex in response to different stimuli.
One of the major challenges in studying RNA-RBP interactions and identifying specific RNA binding proteins is the intricate nature of axonal mRNP complexes (Gumy, Katrukha, Kapitein, & Hoogenraad, 2014; Holt & Bullock, 2009). On one hand, a single RBP can bind multiple transcripts (Donnelly et al., 2013). Hence, the association of a particular mRNA to an RBP can be masked by the presence of other, highly abundant mRNAs that bind to the same RBP. The advantage of the RNA affinity chromatography described herein is the use of a single RNA bait in high quantity so that it can compete with the endogenous transcripts, allowing the detection of various RBPs with a different range of affinities. On the other hand, a specific transcript can be in complex with more than one RBP (Campbell & Wickens, 2015; Pique, Lopez, Foissac, Guigo, & Mendez, 2008; X. Wu et al., 2013), making the identification of direct interactions harder. Hence, it is highly recommended to validate the identified interactions with complementary approaches such as the RIP described here or immunocytochemistry combined with in situ hybridizations for co-localization of the RNA of interest with the specific RBP (Buxbaum, Haimovich, & Singer, 2015). Moreover, as the affinity assay described here cannot distinguish direct from indirect binding, we suggest using RNA affinity chromatography after in vitro translation and/or with the use of recombinant proteins (Tsai et al., 2010) in order to infer whether an interaction is direct or mediated by linker components. In addition, the CLIP methodology (Sugimoto et al., 2012; Ule et al., 2003; Zhang & Darnell, 2011) can determine direct interactions as well as provide information on binding sites.
Other difficulties in studying axonal mRNPs are typically high complexity and low quantity of axonal samples. Moreover, only a fraction of the mRNA of interest may be bound to specific RBPs at a specific point in time and space and thus, identification of RNA-protein interactions are limited by yield. The relative simplicity of the axoplasm preparation method allows working with a large amount of starting material in order to make up for this issue. An additional level of complexity is that some interactions may be constrained to a specific subcellular compartment. Some RNA binding proteins have different roles in the cell body than in the axon (Hornberg & Holt, 2013) and therefore, may interact with different binding partners in each cellular compartment. Working with axoplasm as described here has the appealing advantage of looking at the subcellular compartment relevant for axonal transport. For example, Hanz et al. were able to show differences in axonal importin β1 protein bound to dynein before and after axonal injury using the mentioned axoplasm protocol, despite the fact that this protein is very abundant in cell nuclei (Hanz et al., 2003). Working with whole cell extracts in that case would have completely masked the change after injury. However, it should be considered that the axoplasm preparation protocols described above allow enrichment with axoplasmic components, but do not provide completely pure axonal extract (Rishal et al., 2010). Preparations may contain both glial and non-neuronal contaminants, necessitating care in interpretation of results. If possible, key results can be validated using pure axonal extracts from compartmentalized cultures (described in (Willis & Twiss, 2011)).
An additional aspect of the aforementioned complexity stems from the fact that some interactions are transient or specific to certain physiological conditions. For example, peripheral injury induces increased axonal transport of NMP35 and Neuritin1 (Merianda, Gomes, Yoo, Vuppalanchi, & Twiss, 2013; Merianda, Vuppalanchi, Yoo, Blesch, & Twiss, 2013) and NGF signaling increases localization of Impa1 into axons (Andreassi et al., 2010), processes that are likely regulated by changes in the mRNP complex. Another example for specifically induced mRNA-protein interactions comes from evidence showing that IL-6 and NGF can enhance the formation of eIF4F translation initiation complex on mRNAs to induce local translation in axons of nociceptive neurons (Melemedjian et al., 2010). The conditions used in the suggested RIP and co-IP protocols are in essence native conditions; the mRNP is precipitated as a complex by the specific antibody and the RNA or proteins are released only thereafter. This allows the study of relevant mRNP complexes under various physiological states by conducting pull-downs under different conditions and thus compensating for the transient nature of some interactions.
To conclude, RNA transport and local translation are increasingly being recognized as important processes in axonal physiology. Therefore, analyses of mRNP complexes and their transport is required for understanding diverse aspects of axonal biology. Such analyses are challenging due to the high complexity and limiting quantities of axonal samples, and the intricacy of mRNP complexes. Here we describe a combination of affinity assays that allow the identification and study of mRNPs of specific mRNAs of interest under various physiological conditions.
Acknowledgments
We gratefully acknowledge support from the USA-Israel Binational Science Foundation (2011329, M.F. & J.L.T.), the US Army Medical Research Program (W81XWH-13-1-0308, J.L.T. & M.F.), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation ( J.L.T., M.F., & A.L.B.), the European Research Council (Neurogrowth, M.F.), the Israel Science Foundation (1284/13, M.F.), the National Institutes of Health (R01-NS041596, J.L.T.; P41-GM103481 & S10-OD016229, A.L.B.), and the Howard Hughes Medical Institute (A.L.B.).
References
- Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Saiardi A, Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13(3):291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- Anko ML, Neugebauer KM. RNA-protein interactions in vivo: global gets specific. Trends Biochem Sci. 2012;37(7):255–262. doi: 10.1016/j.tibs.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Aviv T, Lin Z, Ben-Ari G, Smibert CA, Sicheri F. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat Struct Mol Biol. 2006;13(2):168–176. doi: 10.1038/nsmb1053. [DOI] [PubMed] [Google Scholar]
- Ben-Yaakov K, Dagan SY, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, … Fainzilber M. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J. 2012;31(6):1350–1363. doi: 10.1038/emboj.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2015;16(2):95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell ZT, Wickens M. Probing RNA-protein networks: biochemistry meets genomics. Trends Biochem Sci. 2015;40(3):157–164. doi: 10.1016/j.tibs.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, … Hentze MW. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149(6):1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10(2):149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107(4):489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Donnelly CJ, Park M, Spillane M, Yoo S, Pacheco A, Gomes C, … Twiss JL. Axonally synthesized beta-actin and GAP-43 proteins support distinct modes of axonal growth. J Neurosci. 2013;33(8):3311–3322. doi: 10.1523/JNEUROSCI.1722-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche R, Karra D, Bennett KL, Ang FY, Heraud-Farlow JE, Tolino M, … Kiebler MA. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 2013;5(6):1749–1762. doi: 10.1016/j.celrep.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Gomes C, Merianda TT, Lee SJ, Yoo S, Twiss JL. Molecular determinants of the axonal mRNA transcriptome. Dev Neurobiol. 2014;74(3):218–232. doi: 10.1002/dneu.22123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Katrukha EA, Kapitein LC, Hoogenraad CC. New insights into mRNA trafficking in axons. Dev Neurobiol. 2014;74(3):233–244. doi: 10.1002/dneu.22121. [DOI] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, … Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40(6):1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Hengst U, Deglincerti A, Kim HJ, Jeon NL, Jaffrey SR. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat Cell Biol. 2009;11(8):1024–1030. doi: 10.1038/ncb1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Goff SP. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell. 2010;143(3):379–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Bullock SL. Subcellular mRNA Localization in Animal Cells and Why It Matters. Science. 2009;326(5957):1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberg H, Holt C. RNA-binding proteins and translational regulation in axons and growth cones. Front Neurosci. 2013;7:81. doi: 10.3389/fnins.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Stoecklin G. An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins. Nucleic Acids Res. 2014;42(2):e13. doi: 10.1093/nar/gkt956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9(10):1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, … Darnell RB. HITS- CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc Natl Acad Sci U S A. 1996;93(20):10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melemedjian OK, Asiedu MN, Tillu DV, Peebles KA, Yan J, Ertz N, … Price TJ. IL-6- and NGF-induced rapid control of protein synthesis and nociceptive plasticity via convergent signaling to the eIF4F complex. J Neurosci. 2010;30(45):15113–15123. doi: 10.1523/JNEUROSCI.3947-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianda TT, Gomes C, Yoo S, Vuppalanchi D, Twiss JL. Axonal localization of neuritin/CPG15 mRNA in neuronal populations through distinct 5′ and 3′ UTR elements. J Neurosci. 2013;33(34):13735–13742. doi: 10.1523/JNEUROSCI.0962-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianda TT, Vuppalanchi D, Yoo S, Blesch A, Twiss JL. Axonal transport of neural membrane protein 35 mRNA increases axon growth. J Cell Sci. 2013;126(Pt 1):90–102. doi: 10.1242/jcs.107268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milek M, Wyler E, Landthaler M. Transcriptome-wide analysis of protein-RNA interactions using high-throughput sequencing. Semin Cell Dev Biol. 2012;23(2):206–212. doi: 10.1016/j.semcdb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26(2):182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- O'Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516(7530):263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45(5):715–726. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Perry RB, Doron-Mandel E, Iavnilovitch E, Rishal I, Dagan SY, Tsoory M, … Fainzilber M. Subcellular knockout of importin beta1 perturbs axonal retrograde signaling. Neuron. 2012;75(2):294–305. doi: 10.1016/j.neuron.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Anderson R, Dwivedy A, Weinl C, van Horck F, Leung KM, … Holt C. Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron. 2006;49(2):215–228. doi: 10.1016/j.neuron.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132(3):434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- Rishal I, Fainzilber M. Axon-soma communication in neuronal injury. Nat Rev Neurosci. 2014;15(1):32–42. doi: 10.1038/nrn3609. [DOI] [PubMed] [Google Scholar]
- Rishal I, Michaelevski I, Rozenbaum M, Shinder V, Medzihradszky KF, Burlingame AL, Fainzilber M. Axoplasm isolation from peripheral nerve. Dev Neurobiol. 2010;70(2):126–133. doi: 10.1002/dneu.20755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Welshhans K, Wen Z, Yao J, Xu M, Goshima Y, … Bassell GJ. Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local beta-actin synthesis and growth cone turning. J Neurosci. 2010;30(28):9349–9358. doi: 10.1523/JNEUROSCI.0499-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodin B, Gerst JE. A novel mRNA affinity purification technique for the identification of interacting proteins and transcripts in ribonucleoprotein complexes. RNA. 2010;16(11):2277–2290. doi: 10.1261/rna.2091710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Konig J, Hussain S, Zupan B, Curk T, Frye M, Ule J. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 2012;13(8):R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, … Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova- regulated RNA networks in the brain. Science. 2003;302(5648):1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25(2):331–342. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, Twiss JL. Profiling axonal mRNA transport. Methods Mol Biol. 2011;714:335–352. doi: 10.1007/978-1-61779-005-8_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436(7053):1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chesoni S, Rondeau G, Tempesta C, Patel R, Charles S, … Brewer G. Combinatorial mRNA binding by AUF1 and Argonaute 2 controls decay of selected target mRNAs. Nucleic Acids Res. 2013;41(4):2644–2658. doi: 10.1093/nar/gks1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9(10):1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- Yoon BC, Jung H, Dwivedy A, O'Hare CM, Zivraj KH, Holt CE. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;148(4):752–764. doi: 10.1016/j.cell.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, … Fainzilber M. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59(2):241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29(7):607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JQ, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci. 2001;21(23):9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]