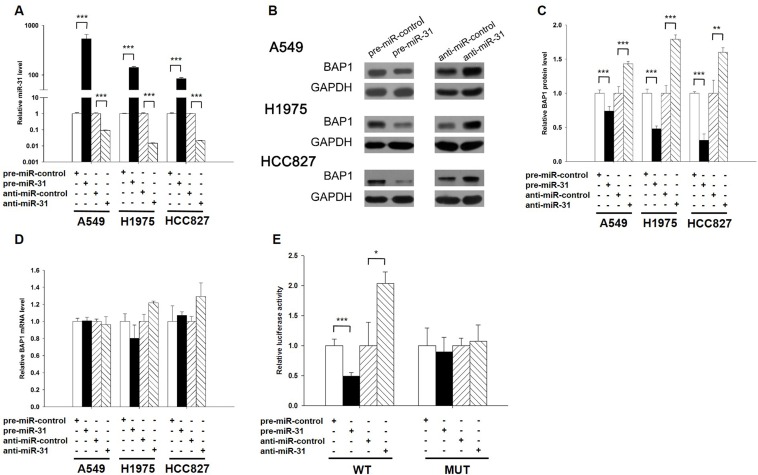
Figure 3. BAP1 is a direct target of miR-31.
A. Quantitative RT-PCR analysis of the expression levels of miR-31 in A549, H1975 and HCC827 cells transfected with equal doses of the miR-31 mimic (pre-miR-31), miR-31 inhibitor (anti-miR-31) or scrambled negative control RNA (pre-miR-control or anti-miR-control). B. and C. Western blotting analysis to detect BAP1 protein levels in A549, H1975 and HCC827 cells transfected with equal doses of the miR-31 mimic, miR-31 inhibitor or scrambled negative control RNA. B: representative image; C. quantitative analysis. D. Quantitative RT-PCR analysis of BAP1 mRNA levels in A549, H1975 and HCC827 cells transfected with equal doses of the miR-31 mimic, miR-31 inhibitor or scrambled negative control RNA. E. Direct recognition of the BAP1 3′-UTR by miR-31. Firefly luciferase reporters containing either wild-type (WT) or mutant (MUT) miR-31 binding sites in the BAP1 3′-UTR were co-transfected into A549 cells with equal doses of the miR-31 mimic, miR-31 inhibitor or scrambled negative control RNA. Twenty-four hours post-transfection, the cells were assayed using a luciferase assay kit. Firefly luciferase values were normalized to β-galactosidase activity, and the results were calculated as the ratio of firefly luciferase activity in the miR-31-transfected cells normalized to the negative control RNA-transfected cells. The results are presented as the mean ± S.D. of three independent experiments. (* p < 0.05; ** p < 0.01; *** p < 0.005).